8T7G
 
 | | Structure of the CK variant of Fab F1 (FabC-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
 | | Structure of the S1 variant of Fab F1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T58
 
 | |
8TS5
 
 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRT
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, monoclinic form | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 2, S1CE variant of Fab C1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRS
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
3ENC
 
 | | Crystal structure of Pyrococcus furiosus PCC1 dimer | | Descriptor: | protein PCC1 | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6304 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
3ENP
 
 | | Crystal structure of human cgi121 | | Descriptor: | TP53RK-binding protein | | Authors: | Haffani, Y.Z, Ceccarelli, D.F, Neculai, D, Mao, D.Y, Sicheri, F. | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
3EN9
 
 | | Structure of the Methanococcus jannaschii KAE1-BUD32 fusion protein | | Descriptor: | HEXATANTALUM DODECABROMIDE, MAGNESIUM ION, O-sialoglycoprotein endopeptidase/protein kinase | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
3ENO
 
 | |
7MM1
 
 | | PTP1B in complex with TCS401 by Native S-SAD at Room Temperature | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Greisman, J.B, Dalton, K.M, Hekstra, D.R. | | Deposit date: | 2021-04-29 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Native SAD phasing at room temperature.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LUO
 
 | | N-terminus of Skp2 bound to Cyclin A | | Descriptor: | S-phase kinase-associated protein 2,Cyclin-A2, Skp2 Motif 1 uncharacterized fragment 1, Skp2 Motif 1 uncharacterized fragment 2 | | Authors: | Kelso, S, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Bipartite binding of the N terminus of Skp2 to cyclin A.
Structure, 29, 2021
|
|
7L84
 
 | |
6B0T
 
 | |
6C16
 
 | |
7UKZ
 
 | |
6BVA
 
 | |
7U3G
 
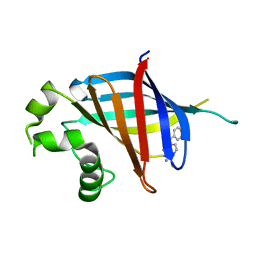 | | GID4 in complex with compound 67 | | Descriptor: | (1R)-1-phenyl-1,2,3,4-tetrahydroisoquinolin-5-amine, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3K
 
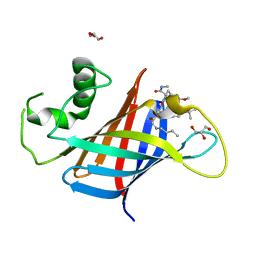 | | GID4 in complex with compound 89 | | Descriptor: | GLYCEROL, Glucose-induced degradation protein 4 homolog, N-butylglycyl-4-tert-butyl-D-phenylalanyl-3-methoxy-N-methyl-L-phenylalaninamide | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3I
 
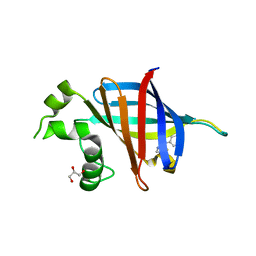 | | GID4 in complex with compound 16 | | Descriptor: | 3-{[(5S)-5-methyl-4,5-dihydro-1,3-thiazol-2-yl]amino}phenol, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3J
 
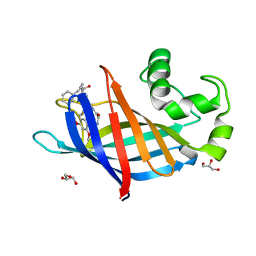 | | GID4 in complex with compound 88 | | Descriptor: | (2S)-2-{[(2S)-2-({N-[(2,4-dimethoxyphenyl)methyl]glycyl}amino)-2-(thiophen-2-yl)acetyl]amino}-N-methyl-4-phenylbutanamide, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3E
 
 | | GID4 in complex with compound 1 | | Descriptor: | Glucose-induced degradation protein 4 homolog, tert-butyl (1S,4S)-2,5-diazabicyclo[2.2.1]heptane-2-carboxylate | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3F
 
 | | GID4 in complex with compound 4 | | Descriptor: | (4R)-4-(4-methoxyphenyl)-4,5,6,7-tetrahydrothieno[3,2-c]pyridine, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3H
 
 | | GID4 in complex with compound 7 | | Descriptor: | (5R)-N-(4-fluorophenyl)-5-methyl-4,5-dihydro-1,3-thiazol-2-amine, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3L
 
 | | GID4 in complex with compound 91 | | Descriptor: | GLYCEROL, Glucose-induced degradation protein 4 homolog, Nalpha-{(2R,4E)-2-[(N-benzylglycyl)amino]-5-phenylpent-4-enoyl}-N,4-dimethyl-L-phenylalaninamide | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
