6R7D
 
 | | Crystal structure of LTC4S in complex with AZ13690257 | | Descriptor: | (1~{S},2~{S})-2-[5-[cyclopropylmethyl(naphthalen-1-yl)amino]-4-methoxy-pyrimidin-2-yl]carbonylcyclopropane-1-carboxylic acid, DODECYL-BETA-D-MALTOSIDE, Leukotriene C4 synthase, ... | | Authors: | Kack, H, Ek, M. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of the Oral Leukotriene C4 Synthase Inhibitor (1S,2S)-2-({5-[(5-Chloro-2,4-difluorophenyl)(2-fluoro-2-methylpropyl)amino]-3-methoxypyrazin-2-yl}carbonyl)cyclopropanecarboxylic Acid (AZD9898) as a New Treatment for Asthma.
J.Med.Chem., 62, 2019
|
|
6SB1
 
 | | Crystal structure of murine perforin-2 P2 domain crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
6SB3
 
 | |
6SB4
 
 | | Crystal structure of murine perforin-2 P2 domain crystal form 2 | | Descriptor: | Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Yu, X, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
6SB5
 
 | |
1GHD
 
 | | Crystal structure of the glutaryl-7-aminocephalosporanic acid acylase by mad phasing | | Descriptor: | GLUTARYL-7-AMINOCEPHALOSPORANIC ACID ACYLASE | | Authors: | Ding, Y, Jiang, W, Mao, X, He, H, Zhang, S, Tang, H, Bartlam, M, Ye, S, Jiang, F, Liu, Y, Zhao, G, Rao, Z. | | Deposit date: | 2000-12-07 | | Release date: | 2003-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Affinity alkylation of the Trp-B4 residue of the beta -subunit of the glutaryl 7-aminocephalosporanic acid acylase of Pseudomonas sp. 130.
J.Biol.Chem., 277, 2002
|
|
7C53
 
 | |
5B6G
 
 | | Protein-protein interaction | | Descriptor: | Adenomatous polyposis coli protein, GLYCEROL, PHQ-ALA-GLY-GLU-ALA-XYC-TYR-GLU, ... | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
1J4J
 
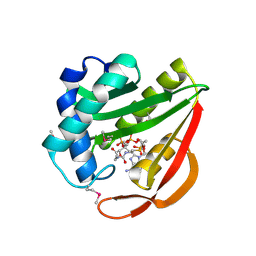 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
4N8R
 
 | | Crystal structure of RXRa LBD complexed with a synthetic modulator K-8008 | | Descriptor: | 5-(2-{(1Z)-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
4N5G
 
 | | Crystal Structure of RXRa LBD complexed with a synthetic modulator K8012 | | Descriptor: | 5-(2-{(1Z)-5-fluoro-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
7FEM
 
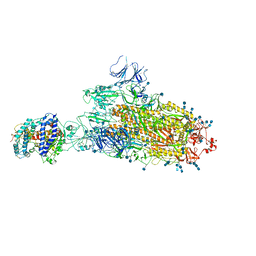 | | SARS-CoV-2 B.1.1.7 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wen, Z.L, Zhu, Y, Sun, F. | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure-based evidence for the enhanced transmissibility of the dominant SARS-CoV-2 B.1.1.7 variant (Alpha).
Cell Discov, 7, 2021
|
|
7FET
 
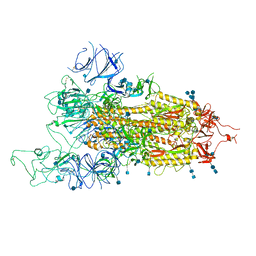 | | SARS-CoV-2 B.1.1.7 Spike Glycoprotein trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wen, Z.L, Zhu, Y, Sun, F. | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure-based evidence for the enhanced transmissibility of the dominant SARS-CoV-2 B.1.1.7 variant (Alpha).
Cell Discov, 7, 2021
|
|
7M3U
 
 | |
6ZHC
 
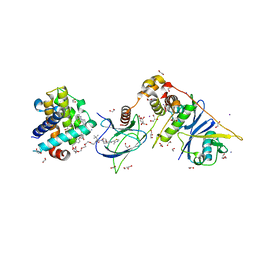 | | PROTAC6 mediated complex of VHL:EloB:EloC and Bcl-xL | | Descriptor: | 1,2-ETHANEDIOL, 2-[8-(1,3-benzothiazol-2-ylcarbamoyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]-5-[3-[4-[3-[2-[2-[2-[2-[2-[3-[[(2~{S})-3,3-dimethyl-1-[(2~{S},4~{R})-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-3-oxidanylidene-propoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]prop-1-ynyl]phenoxy]propyl]-1,3-thiazole-4-carboxylic acid, Bcl-2-like protein 1, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into PROTAC-Mediated Degradation of Bcl-xL.
Acs Chem.Biol., 15, 2020
|
|
7MMY
 
 | |
4FVT
 
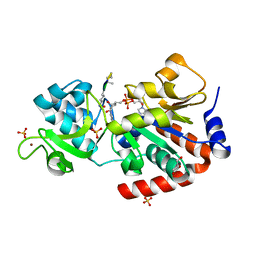 | | Human SIRT3 bound to Ac-ACS peptide and Carba-NAD | | Descriptor: | Acetylated ACS2 peptide, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Dai, H. | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-15 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Synthesis of Carba-NAD and the Structures of Its Ternary Complexes with SIRT3 and SIRT5.
J.Org.Chem., 77, 2012
|
|
4G1C
 
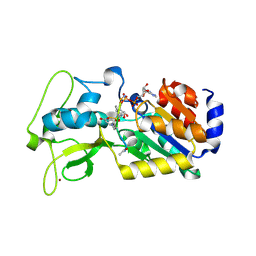 | | Human SIRT5 bound to Succ-IDH2 and Carba-NAD | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-dependent protein deacylase sirtuin-5, mitochondrial, ... | | Authors: | Dai, H. | | Deposit date: | 2012-07-10 | | Release date: | 2012-08-15 | | Last modified: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Synthesis of Carba-NAD and the Structures of Its Ternary Complexes with SIRT3 and SIRT5.
J.Org.Chem., 77, 2012
|
|
4G1N
 
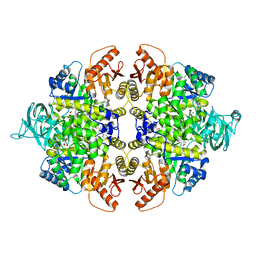 | | PKM2 in complex with an activator | | Descriptor: | MAGNESIUM ION, N-(4-{[4-(pyrazin-2-yl)piperazin-1-yl]carbonyl}phenyl)quinoline-8-sulfonamide, OXALATE ION, ... | | Authors: | Kung, C, Hixon, J, Dang, L, DeLaBarre, B, Qian, K.C. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small Molecule Activation of PKM2 in Cancer Cells Induces Serine Auxotrophy.
Chem.Biol., 19, 2012
|
|
1MOL
 
 | |
4JA8
 
 | | Complex of Mitochondrial Isocitrate Dehydrogenase R140Q Mutant with AGI-6780 Inhibitor | | Descriptor: | 1-[5-(cyclopropylsulfamoyl)-2-thiophen-3-yl-phenyl]-3-[3-(trifluoromethyl)phenyl]urea, CALCIUM ION, GLYCEROL, ... | | Authors: | Wei, W, Chen, L, Wu, M, Jiang, F, Travins, J, Qian, K, DeLaBarre, B. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation.
Science, 340, 2013
|
|
7TEO
 
 | | Cryo-EM structure of the 20S Alpha 3 Deletion proteasome core particle in complex with FUB1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-4, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Schnell, H.M, Hanna, J. | | Deposit date: | 2022-01-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Yeast PI31 inhibits the proteasome by a direct multisite mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TEJ
 
 | | Cryo-EM structure of the 20S Alpha 3 Deletion proteasome core particle | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-4, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Schnell, H.M, Hanna, J. | | Deposit date: | 2022-01-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Yeast PI31 inhibits the proteasome by a direct multisite mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5IZ8
 
 | | Protein-protein interaction | | Descriptor: | ACE-ALA-GLY-GLU-ALA-LEU-ALA-ASP-NH2, Adenomatous polyposis coli protein, TRIETHYLENE GLYCOL | | Authors: | Zhao, Y, Jiang, H, Yang, X, Jiang, F, Song, K, Zhang, J. | | Deposit date: | 2016-03-25 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Nat. Chem. Biol., 13, 2017
|
|
5J3F
 
 | | NMR solution structure of [Rp, Rp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(RSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(RSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
