8EX4
 
 | | Human S1P transporter Spns2 in an inward-facing open conformation (state 1) | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Ahmed, S, Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8EX7
 
 | |
8EX8
 
 | |
1Q1M
 
 | | A Highly Efficient Approach to a Selective and Cell Active PTP1B inhibitors | | Descriptor: | 5-{2-FLUORO-5-[3-(3-HYDROXY-2-METHOXYCARBONYL-PHENOXY)-PROPENYL]-PHENYL}-ISOXAZOLE-3-CARBOXYLIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Xin, Z, Pei, Z, Hajduk, P.J, Abad-Zapatero, C, Hutchins, C.W, Zhao, H, Lubben, T.H, Ballaron, S.J, Haasch, D.L, Kaszubska, W, Rondinone, C.M, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-07-22 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment screening and assembly: a highly efficient approach to a selective and cell active protein tyrosine phosphatase 1B inhibitor.
J.Med.Chem., 46, 2003
|
|
1NVQ
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/UCN-01 | | Descriptor: | 7-HYDROXYSTAUROSPORINE, Peptide ASVSA, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NVR
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/Staurosporine | | Descriptor: | Peptide ASVSA, STAUROSPORINE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
1NVS
 
 | | The Complex Structure Of Checkpoint Kinase Chk1/SB218078 | | Descriptor: | Peptide ASVSA, REL-(9R,12S)-9,10,11,12-TETRAHYDRO-9,12-EPOXY-1H-DIINDOLO[1,2,3-FG:3',2',1'-KL]PYRROLO[3,4-I][1,6]BENZODIAZOCINE-1,3(2H)-DIONE, SULFATE ION, ... | | Authors: | Zhao, B, Bower, M.J, McDevitt, P.J, Zhao, H, Davis, S.T, Johanson, K.O, Green, S.M, Concha, N.O, Zhou, B.B. | | Deposit date: | 2003-02-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Chk1 Inhibition by UCN-01
J.Biol.Chem., 277, 2002
|
|
7TZL
 
 | | The DH dehydratase domain of AlnB | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Swain, K, Blackson, W, Wang, B, Zhao, H, Nannenga, B.L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The programming of alpha,beta-polyene biosynthesis by a bacterial iterative type I polyketide synthase
To Be Published
|
|
6M6E
 
 | |
6M6F
 
 | |
5VGT
 
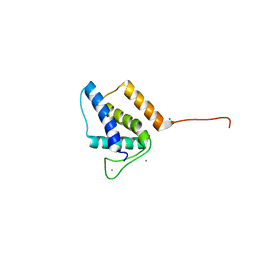 | | X-ray structure of bacteriophage Sf6 tail adaptor protein gp7 | | Descriptor: | CALCIUM ION, Gene 7 protein, MAGNESIUM ION | | Authors: | Tang, L, Liang, L, Zhao, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7YN9
 
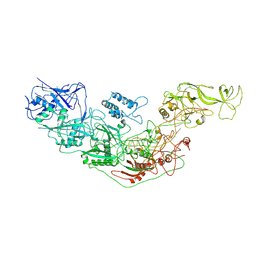 | | Cryo-EM structure of Cas7-11-crRNA binary complex | | Descriptor: | CRISPR-associated RAMP family protein, crRNA | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNC
 
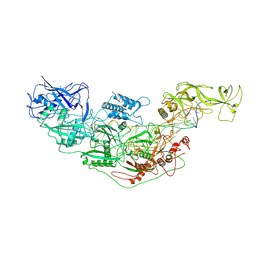 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-3 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-3 (28-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNA
 
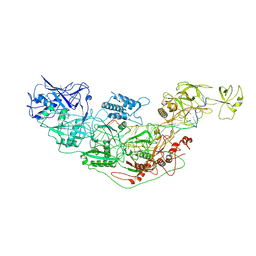 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-1 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-1 (25-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YNB
 
 | | Cryo-EM structure of Cas7-11-crRNA bound to target RNA-2 | | Descriptor: | CRISPR-associated RAMP family protein, Target RNA-2 (28-MER), crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
7YND
 
 | | Cryo-EM structure of Cas7-11-crRNA-Csx29 ternary complex | | Descriptor: | CHAT domain-containing protein, CRISPR-associated RAMP family protein, crRNA (38-MER) | | Authors: | Huo, Y, Dong, Q, Zhao, H, Jiang, T. | | Deposit date: | 2022-07-30 | | Release date: | 2023-02-01 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure and protease activity of the type III-E CRISPR-Cas effector.
Nat Microbiol, 8, 2023
|
|
3WRS
 
 | |
3WRQ
 
 | |
1MRY
 
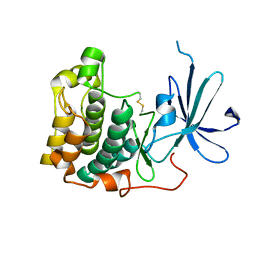 | | crystal structure of an inactive akt2 kinase domain | | Descriptor: | RAC-beta serine/threonine kinase | | Authors: | Huang, X, Begley, M, Morgenstern, K.A, Gu, Y, Rose, P, Zhao, H, Zhu, X. | | Deposit date: | 2002-09-18 | | Release date: | 2003-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an inactive akt2 kinase domain
Structure, 11, 2003
|
|
1MRV
 
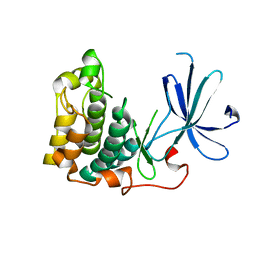 | | crystal structure of an inactive Akt2 kinase domain | | Descriptor: | RAC-beta serine/threonine kinase | | Authors: | Huang, X, Begley, M, Morgenstern, K.A, Gu, Y, Rose, P, Zhao, H, Zhu, X. | | Deposit date: | 2002-09-18 | | Release date: | 2003-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an inactive akt2 kinase domain
Structure, 11, 2003
|
|
1QPD
 
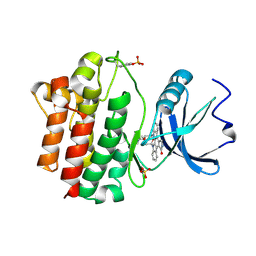 | | STRUCTURAL ANALYSIS OF THE LYMPHOCYTE-SPECIFIC KINASE LCK IN COMPLEX WITH NON-SELECTIVE AND SRC FAMILY SELECTIVE KINASE INHIBITORS | | Descriptor: | LCK KINASE, STAUROSPORINE, SULFATE ION | | Authors: | Zhu, X, Kim, J.L, Rose, P.E, Stover, D.R, Toledo, L.M, Zhao, H, Morgenstern, K.A. | | Deposit date: | 1999-05-24 | | Release date: | 2000-05-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors.
Structure Fold.Des., 7, 1999
|
|
7K0K
 
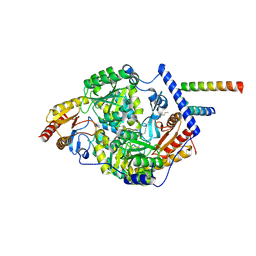 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa, 3KS-bound | | Descriptor: | 3-Dehydrosphinganine, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0O
 
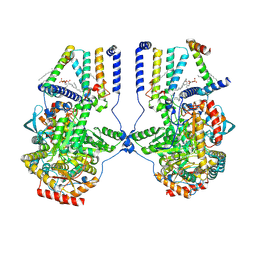 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 3 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0N
 
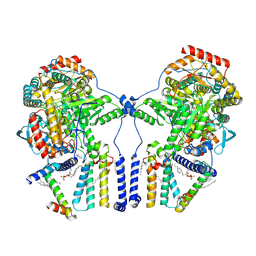 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa/ORMDL3, class 2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K0J
 
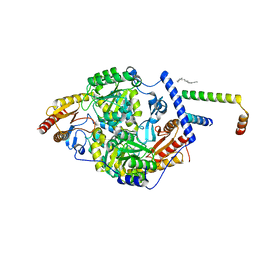 | | Human serine palmitoyltransferase complex SPTLC1/SPLTC2/ssSPTa protomer | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Wang, Y, Niu, Y, Zhang, Z, Zhao, H, Myasnikov, A, Kalathur, R, Lee, C.H. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the regulation of human serine palmitoyltransferase complexes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
