8P09
 
 | | 48S late-stage initiation complex with non methylated mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Guca, E, Lima, L.H.F, Boissier, F, Hashem, Y. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | N 6 -methyladenosine in 5' UTR does not promote translation initiation.
Mol.Cell, 84, 2024
|
|
8P03
 
 | | 48S late-stage initiation complex with m6A mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Guca, E, Lima, L.H.F, Boissier, F, Hashem, Y. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | N 6 -methyladenosine in 5' UTR does not promote translation initiation.
Mol.Cell, 84, 2024
|
|
6CHZ
 
 | | Estrogen Receptor Alpha Y537S bound to antagonist H3B-9224. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, Estrogen receptor | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
6CHW
 
 | | Estrogen Receptor Alpha Y537S covalently bound to antagonist H3B-5942. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
8OXW
 
 | |
8OXX
 
 | |
8OXY
 
 | |
8OXV
 
 | |
2MI1
 
 | |
6DDJ
 
 | | Crystal Structure of the human BRD2 BD2 bromodimain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
6DDI
 
 | | Crystal Structure of the human BRD2 BD1 bromodomain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
6YFY
 
 | | Solid-state NMR structure of the D-Arg4,L10-teixobactin - Lipid II complex in lipid bilayers. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, 3-methylbut-2-en-1-ol, D-Arg4,Leu10-Teixobactin, ... | | Authors: | Weingarth, M.H, Shukla, R. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-08-07 | | Method: | SOLID-STATE NMR | | Cite: | Mode of action of teixobactins in cellular membranes.
Nat Commun, 11, 2020
|
|
6WWB
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the compound 3b | | Descriptor: | 1,2-ETHANEDIOL, 2-((S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)-N-((1-(4-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)acetamido)butyl)-1H-1,2,3-triazol-4-yl)methyl)acetamide, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Phenyl-Glutarimides: Alternative Cereblon Binders for the Design of PROTACs.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6SDG
 
 | |
8FTQ
 
 | | Crystal structure of hRpn13 Pru domain in complex with Ubiquitin and XL44 | | Descriptor: | N-(3-{[(3R)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]methyl}phenyl)-4-methoxybenzamide, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Walters, K.J, Lu, X, Chandravanshi, M. | | Deposit date: | 2023-01-13 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based designed small molecule depletes hRpn13 Pru and a select group of KEN box proteins.
Nat Commun, 15, 2024
|
|
5DNI
 
 | | Crystal structure of Methanocaldococcus jannaschii Fumarate hydratase beta subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jayaraman, V, Kunala, J, Balaram, H. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Revisiting the Burden Borne by Fumarase: Enzymatic Hydration of an Olefin.
Biochemistry, 62, 2023
|
|
6SC4
 
 | | Gamma-Carbonic Anhydrase from the Haloarchaeon Halobacterium sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Vogler, M, Karan, R, Renn, D, Vancea, A, Vielberg, V.-T, Groetzinger, S.W, DasSarma, P, Das Sarma, S, Eppinger, J, Groll, M, Rueping, M. | | Deposit date: | 2019-07-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Active Site Engineering of a Halophilic gamma-Carbonic Anhydrase.
Front Microbiol, 11, 2020
|
|
7DK1
 
 | | Crystal structure of Zinc bound SARS-CoV-2 main protease | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Sonkar, K.S, Panchariya, L, Kuila, S, Khan, W.A, Arockiasamy, A. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Zinc 2+ ion inhibits SARS-CoV-2 main protease and viral replication in vitro.
Chem.Commun.(Camb.), 57, 2021
|
|
2IVX
 
 | | Crystal structure of human cyclin T2 at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CYCLIN-T2 | | Authors: | Debreczeni, J.E, Bullock, A.N, Fedorov, O, Savitsky, P, Berridge, G, Das, S, Pike, A.C.W, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Gorrec, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation.
EMBO J., 27, 2008
|
|
2JJD
 
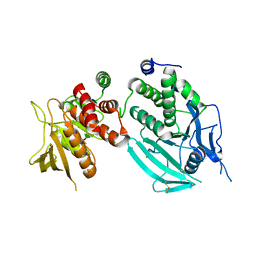 | | Protein Tyrosine Phosphatase, Receptor Type, E isoform | | Descriptor: | CHLORIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE EPSILON | | Authors: | Elkins, J.M, Ugochukwu, E, Alfano, I, Barr, A.J, Bunkoczi, G, King, O.N.F, Filippakopoulos, P, Savitsky, P, Salah, E, Pike, A, Johansson, C, Das, S, Burgess-Brown, N.A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
7FDS
 
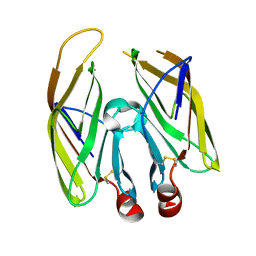 | | High resolution crystal structure of LpqH from Mycobacterium tuberculosis | | Descriptor: | Lipoprotein LpqH | | Authors: | Kundapura, S.V, Chatterjee, S, Samanta, D, Ramagopal, U.A. | | Deposit date: | 2021-07-17 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.258 Å) | | Cite: | High-resolution crystal structure of LpqH, an immunomodulatory surface lipoprotein of Mycobacterium tuberculosis reveals a distinct fold and a conserved cleft on its surface.
Int.J.Biol.Macromol., 210, 2022
|
|
7WVL
 
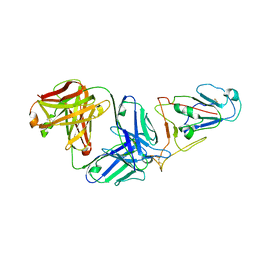 | |
7KXI
 
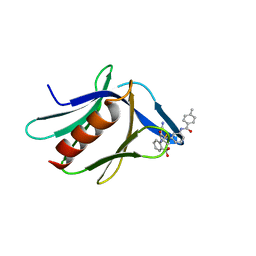 | | Structure of XL5-ligated hRpn13 Pru domain | | Descriptor: | 2-{[(2S)-2-cyano-3-{3-[(4-methylbenzene-1-carbonyl)amino]phenyl}propanoyl]amino}benzoic acid, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-29 | | Method: | SOLUTION NMR | | Cite: | Structure-guided bifunctional molecules hit a DEUBAD-lacking hRpn13 species upregulated in multiple myeloma.
Nat Commun, 12, 2021
|
|
8U7Y
 
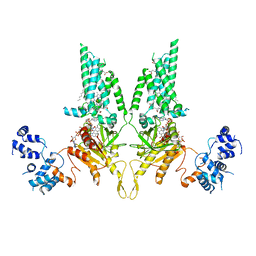 | | Structural Basis of Human NOX5 Activation | | Descriptor: | DECANE, DODECANE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cui, C, Jiang, M, Sun, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural basis of human NOX5 activation.
Nat Commun, 15, 2024
|
|
7D97
 
 | |
