4K3Y
 
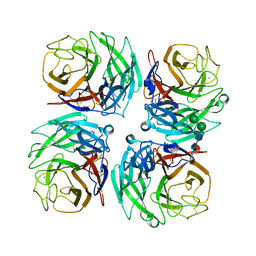 | |
4GZS
 
 | | N2 neuraminidase D151G mutant of a/Tanzania/205/2010 H3N2 in complex with hepes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
4GDI
 
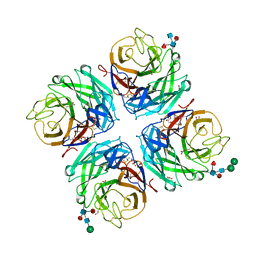 | | A subtype N10 neuraminidase-like protein of A/little yellow-shouldered bat/Guatemala/164/2009 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-07-31 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GXX
 
 | | Crystal structure of the "avianized" 1918 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2012-09-04 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Influenza Human Monoclonal Antibody 1F1 Interacts with Three Major Antigenic Sites and Residues Mediating Human Receptor Specificity in H1N1 Viruses.
Plos Pathog., 8, 2012
|
|
4GZO
 
 | | N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with hepes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
4GZX
 
 | |
4KY2
 
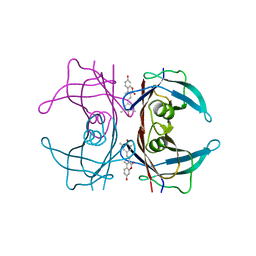 | | Transthyretin in complex with the fluorescent folding sensor (E)-7-hydroxy-3-(4-hydroxy-3,5-dimethylstyryl)-4-methyl-2H-chromen-2-one | | Descriptor: | 7-hydroxy-3-[(E)-2-(4-hydroxy-3,5-dimethylphenyl)ethenyl]-4-methyl-2H-chromen-2-one, Transthyretin | | Authors: | Connelly, S, Wilson, I.A, Choi, S. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Bifunctional coumarin derivatives that inhibit transthyretin amyloidogenesis and serve as fluorescent transthyretin folding sensors.
Chem.Commun.(Camb.), 49, 2013
|
|
4GXU
 
 | |
4JY6
 
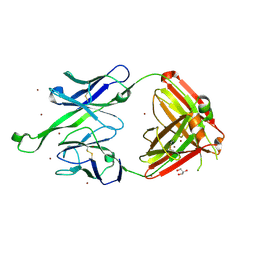 | | Crystal structure of human Fab PGT123, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGT123 heavy chain, PGT123 light chain, ... | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans.
Plos Pathog., 9, 2013
|
|
4DGY
 
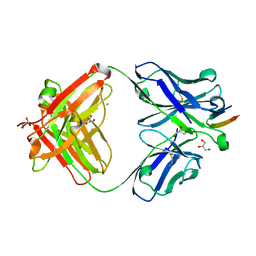 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody HCV1, C2 form | | Descriptor: | CHLORIDE ION, E2 peptide, GLYCEROL, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2012-01-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HFU
 
 | | Crystal structure of Fab 8M2 in complex with a H2N2 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 8M2 heavy chain, Fab 8M2 light chain, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GZQ
 
 | |
4GZT
 
 | | N2 neuraminidase D151G mutant of A/Tanzania/205/2010 H3N2 in complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
4GZP
 
 | | N2 Neuraminidase of A/Tanzania/205/2010 H3N2 in complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
4GZW
 
 | | N2 neuraminidase D151G mutant of A/Tanzania/205/2010 H3N2 in complex with avian sialic acid receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
4L1T
 
 | |
4L1S
 
 | |
1I7Z
 
 | | ANTIBODY GNC92H2 BOUND TO LIGAND | | Descriptor: | CHIMERA OF IG GAMMA-1 CHAIN: HUMAN CONSTANT REGION AND MOUSE VARIABLE REGION, CHIMERA OF IG KAPPA CHAIN: HUMAN CONSTANT REGION AND MOUSE VARIABLE REGION, COCAINE | | Authors: | Larsen, N.A, Wilson, I.A. | | Deposit date: | 2001-03-12 | | Release date: | 2001-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a cocaine-binding antibody.
J.Mol.Biol., 311, 2001
|
|
8E1P
 
 | | Crystal structure of BG505 SOSIP.v4.1-GT1.2 trimer in complex with gl-PGV20 and PGT124 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505-SOSIP.v4.1-GT1.2gp120, ... | | Authors: | Sarkar, A, Kumar, S, Wilson, I.A. | | Deposit date: | 2022-08-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.82 Å) | | Cite: | Germline-targeting HIV-1 Env vaccination induces VRC01-class antibodies with rare insertions.
Cell Rep Med, 4, 2023
|
|
1HQ8
 
 | | CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A | | Descriptor: | NKG2-D | | Authors: | Wolan, D.W, Teyton, L, Rudolph, M.G, Villmow, B, Bauer, S, Busch, D.H, Wilson, I.A. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A.
Nat.Immunol., 2, 2001
|
|
1JCL
 
 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
1JU4
 
 | | BACTERIAL COCAINE ESTERASE COMPLEX WITH PRODUCT | | Descriptor: | BENZOIC ACID, cocaine esterase | | Authors: | Larsen, N.A, Turner, J.M, Stevens, J, Rosser, S.J, Basran, A, Lerner, R.A, Bruce, N.C, Wilson, I.A. | | Deposit date: | 2001-08-23 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of a bacterial cocaine esterase.
Nat.Struct.Biol., 9, 2002
|
|
1I9E
 
 | | TCR DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOTOXIC TCELL VALPHA DOMAIN | | Authors: | Rudolph, M.G, Huang, M, Teyton, L, Wilson, I.A. | | Deposit date: | 2001-03-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an isolated V(alpha) domain of the 2C T-cell receptor.
J.Mol.Biol., 314, 2001
|
|
1HZH
 
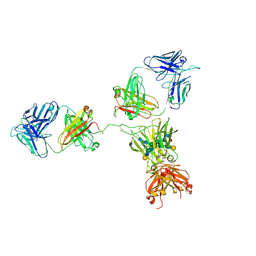 | | CRYSTAL STRUCTURE OF THE INTACT HUMAN IGG B12 WITH BROAD AND POTENT ACTIVITY AGAINST PRIMARY HIV-1 ISOLATES: A TEMPLATE FOR HIV VACCINE DESIGN | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN,Uncharacterized protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Burton, D.R, Wilson, I.A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design.
Science, 293, 2001
|
|
8FO3
 
 | |
