5CN5
 
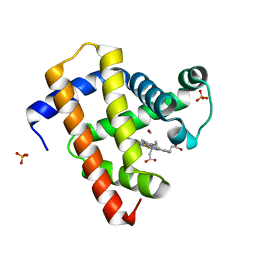 | | Ultrafast dynamics in myoglobin: 0 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5BY6
 
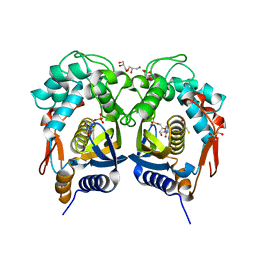 | | Crystal structure of Trichinella spiralis thymidylate synthase complexed with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W, Fraczyk, T, Wilk, P, Rode, W. | | Deposit date: | 2015-06-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
3VXQ
 
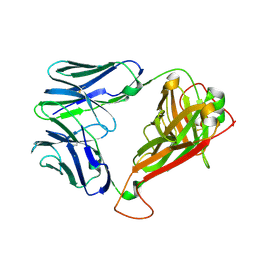 | | H27-14 TCR specific for HLA-A24-Nef134-10 | | Descriptor: | H27-14 TCR alpha chain, H27-14 TCR beta chain | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
5C5V
 
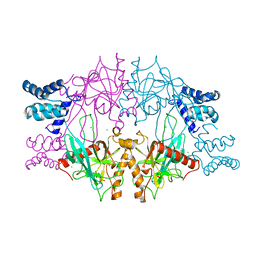 | | Recombinant Inorganic Pyrophosphatase from T brucei brucei | | Descriptor: | 1,2-ETHANEDIOL, Acidocalcisomal pyrophosphatase, BROMIDE ION, ... | | Authors: | Jamwal, A, Yogavel, M, Sharma, A. | | Deposit date: | 2015-06-22 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Highlights of Vacuolar Soluble Protein 1 from Pathogen Trypanosoma brucei brucei
J.Biol.Chem., 290, 2015
|
|
7AYM
 
 | | Structure of DDR2 Kinase domain in complex with IBZ3 | | Descriptor: | Discoidin domain-containing receptor 2,Epithelial discoidin domain-containing receptor 1, N-[5-({[(3-fluorophenyl)carbamoyl]amino}methyl)-2-methylphenyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Nawrotek, A, Talagas, A, Vuillard, L.M, Miallau, L. | | Deposit date: | 2020-11-12 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of DDR2 Kinase domain in complex with IBZ3
To Be Published
|
|
4XUA
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with E11919 BAZ2-ICR analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-4-phenyl-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
8CGL
 
 | |
7AMD
 
 | | In situ assembly of choline acetyltransferase ligands by a hydrothiolation reaction reveals key determinants for inhibitor design | | Descriptor: | Choline O-acetyltransferase, SODIUM ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-2,2-dimethyl-4-[[3-[2-[(1~{R})-2-(1-methylpyridin-4-yl)-1-naphthalen-1-yl-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Allgardsson, A, Ekstrom, F.J, Wiktelius, D, Bergstrom, T, Hoster, N, Akfur, C, Forsgren, N, Lejon, C, Hedenstrom, M, Linusson, A. | | Deposit date: | 2020-10-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | In Situ Assembly of Choline Acetyltransferase Ligands by a Hydrothiolation Reaction Reveals Key Determinants for Inhibitor Design.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8PYW
 
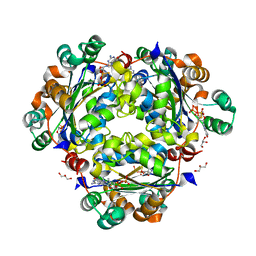 | | Crystal structure of the human Nucleoside-diphosphate kinase B domain bound to compound diphosphate form of AT-9052-Sp. | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [[(2R,3R,4R,5R)-5-(2-azanyl-6-oxidanylidene-1H-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methoxy-sulfanyl-phosphoryl] dihydrogen phosphate | | Authors: | Feracci, M, Chazot, A. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | An exonuclease-resistant chain-terminating nucleotide analogue targeting the SARS-CoV-2 replicase complex.
Nucleic Acids Res., 52, 2024
|
|
8BY2
 
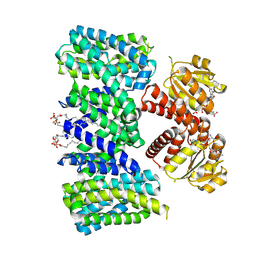 | | Structure of the K+/H+ exchanger KefC with GSH. | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE MONOPHOSPHATE, GLUTATHIONE, ... | | Authors: | Gulati, A, Drew, D. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structure and mechanism of the K + /H + exchanger KefC.
Nat Commun, 15, 2024
|
|
7AOI
 
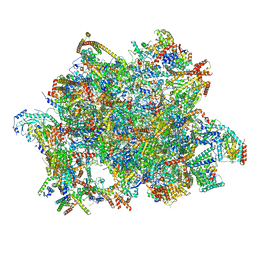 | | Trypanosoma brucei mitochondrial ribosome large subunit assembly intermediate | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Tobiasson, V, Gahura, O, Aibara, S, Baradaran, R, Zikova, A, Amunts, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit.
Embo J., 40, 2021
|
|
5J7G
 
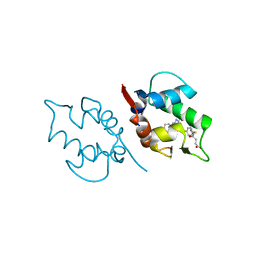 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | Descriptor: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
7NPM
 
 | |
8BZL
 
 | | Human 20S Proteasome in complex with peptide activator peptide BLM42 | | Descriptor: | ARG-SER-TYR-TYR-SER, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Henneberg, F, Chari, A, Jankowska, E, Witkowska, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Peptidic, Blm10-based activators of human 20S proteasome in vitro and in cellulo enhance degradation of proteins connected with neurodegeneration.
To Be Published
|
|
5MPK
 
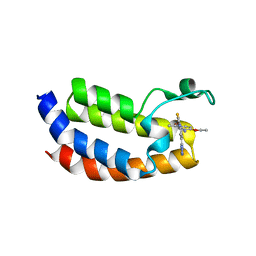 | | Crystal structure of CREBBP bromodomain complexed with DK19 | | Descriptor: | CREB-binding protein, ~{N}-(5-ethanoyl-2-ethoxy-phenyl)-3-(2~{H}-1,2,3,4-tetrazol-5-yl)-5-(1,3-thiazol-4-yl)benzamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
4Y1I
 
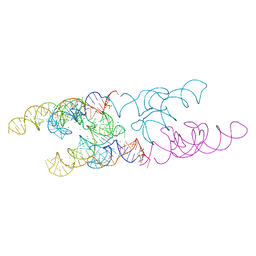 | | Lactococcus lactis yybP-ykoY Mn riboswitch bound to Mn2+ | | Descriptor: | BARIUM ION, GUANOSINE-5'-TRIPHOSPHATE, Lactococcus lactis yybP-ykoY riboswitch, ... | | Authors: | Price, I.R, Ke, A. | | Deposit date: | 2015-02-07 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mn(2+)-Sensing Mechanisms of yybP-ykoY Orphan Riboswitches.
Mol.Cell, 57, 2015
|
|
4ETC
 
 | | Lysozyme, room temperature, 24 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
3O1W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form III | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
5J72
 
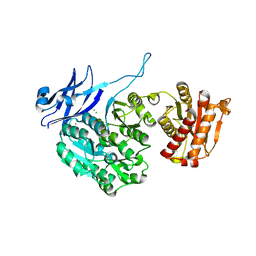 | | Cwp6 from Clostridium difficile | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
5BVK
 
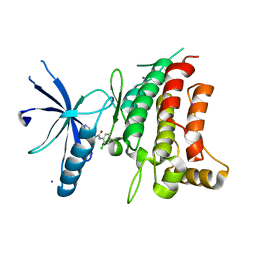 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | 1-(2-chlorophenyl)-3-(pyridin-3-ylmethyl)urea, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5BVO
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(5-{(1S)-1-[(5-fluoro-1,3-benzoxazol-2-yl)amino]ethyl}-2-methylphenyl)imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8C89
 
 | | SARS-CoV-2 spike in complex with the 17T2 neutralizing antibody Fab fragment (local refinement of RBD and Fab) | | Descriptor: | 17T2 Fab heavy chain, 17T2 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Modrego, A, Carlero, D, Bueno-Carrasco, M.T, Santiago, C, Carolis, C, Arranz, R, Blanco, J, Magri, G. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.41 Å) | | Cite: | A monoclonal antibody targeting a large surface of the receptor binding motif shows pan-neutralizing SARS-CoV-2 activity.
Nat Commun, 15, 2024
|
|
8C3B
 
 | | X-ray structure of RNase A upon reaction with a Ruthenium(II)-arene Complexed with Glycosylated Carbene Ligands (5) | | Descriptor: | (1,3-dimethyl-2~{H}-imidazol-2-yl)-oxidanyl-oxidanylidene-ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tetrakis(oxidanyl)ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tris(oxidanyl)ruthenium, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Ruthenium(II)–Arene Complexes with Glycosylated NHC-Carbene Co-Ligands: Synthesis, Hydrolytic Behavior, and Binding to Biological Molecules
Organometallics, 42, 2023
|
|
8C39
 
 | | X-ray structure of HEWL upon reaction with a Ruthenium(II)-arene Complexed with Glycosylated Carbene Ligands (5) | | Descriptor: | CHLORIDE ION, Lysozyme, NITRATE ION, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ruthenium(II)–Arene Complexes with Glycosylated NHC-Carbene Co-Ligands: Synthesis, Hydrolytic Behavior, and Binding to Biological Molecules
Organometallics, 42, 2023
|
|
