1GAX
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS VALYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(VAL) AND VALYL-ADENYLATE ANALOGUE | | Descriptor: | N-[VALINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, TRNA(VAL), VALYL-TRNA SYNTHETASE, ... | | Authors: | Fukai, S, Nureki, O, Sekine, S, Shimada, A, Tao, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-06-23 | | Release date: | 2000-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNA(Val) and valyl-tRNA synthetase.
Cell(Cambridge,Mass.), 103, 2000
|
|
1IVS
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS VALYL-TRNA SYNTHETASE COMPLEXED WITH TRNA(VAL) AND VALYL-ADENYLATE ANALOGUE | | Descriptor: | N-[VALINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, Valyl-tRNA synthetase, tRNA (Val) | | Authors: | Fukai, S, Nureki, O, Sekine, S.-I, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-29 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase
RNA, 9, 2003
|
|
1UAD
 
 | | Crystal structure of the RalA-GppNHp-Sec5 Ral-binding domain complex | | Descriptor: | Exocyst complex component Sec5, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Fukai, S, Matern, H.T, Scheller, R.H, Brunger, A.T. | | Deposit date: | 2003-03-09 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the interaction between RalA and Sec5, a subunit of the Sec6/8 complex
EMBO J., 22, 2003
|
|
1IYW
 
 | | Preliminary Structure of Thermus thermophilus Ligand-Free Valyl-tRNA Synthetase | | Descriptor: | Valyl-tRNA Synthetase | | Authors: | Fukai, S, Nureki, O, Sekine, S, Shimada, A, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-09-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Mechanism of molecular interactions for tRNA(Val) recognition by valyl-tRNA synthetase
RNA, 9, 2003
|
|
2E7S
 
 | |
8J6N
 
 | | Crystal structure of Cystathionine gamma-lyase in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, Cystathionine gamma-lyase, GLYCEROL, ... | | Authors: | Hibi, R, Toma-Fukai, S, Shimizu, T, Hanaoka, K. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a cystathionine gamma-lyase (CSE) selective inhibitor targeting active-site pyridoxal 5'-phosphate (PLP) via Schiff base formation.
Sci Rep, 13, 2023
|
|
4UBQ
 
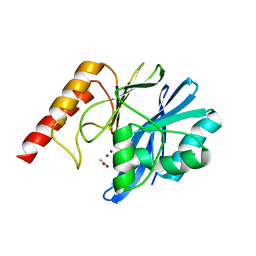 | | Crystal Structure of IMP-2 Metallo-beta-Lactamase from Acinetobacter spp. | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Yamaguchi, Y, Matsueda, S, Matsunaga, K, Takashio, N, Toma-Fukai, S, Yamagata, Y, Shibata, N, Wachino, J, Shibayama, K, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Biol.Pharm.Bull., 38, 2015
|
|
5DST
 
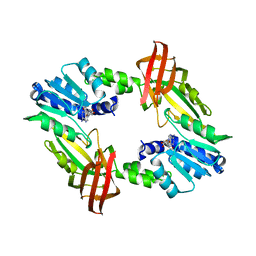 | |
5XIT
 
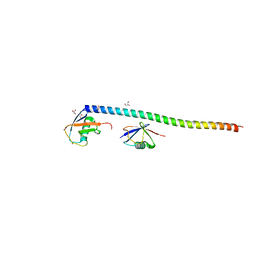 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form II | | Descriptor: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XIS
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form I | | Descriptor: | E3 ubiquitin-protein ligase RNF168, MAGNESIUM ION, Ubiquitin-40S ribosomal protein S27a, ... | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XWU
 
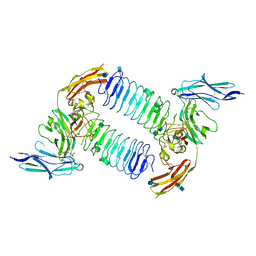 | | Crystal structure of PTPdelta Ig1-Ig3 in complex with SALM2 LRR-Ig | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
4WU5
 
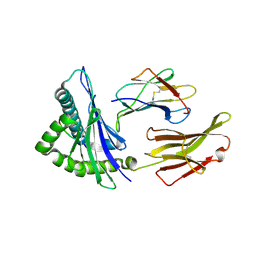 | | HLA-A24 in complex with HIV-1 Nef134-8(wt) | | Descriptor: | 8-Mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Yamagata, A, Fukai, S, Iwamoto, A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Nef134-8(wt) epitope presented by HLA-A24
To Be Published
|
|
4WU7
 
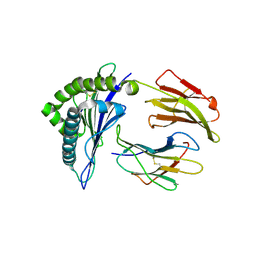 | | HLA-A24 in complex with HIV-1 Nef134-8(2F) | | Descriptor: | 8-Mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Yamagata, A, Fukai, S, Iwamoto, A. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of Nef134-8 epitope presented by HLA-A24
To Be Published
|
|
7EXE
 
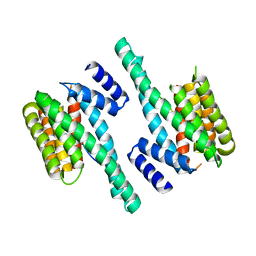 | |
5WQK
 
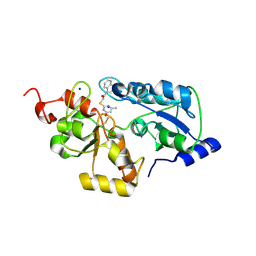 | |
5WQJ
 
 | |
7W86
 
 | | Crystal structure of the DYW domain of DYW1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Sawada, Y, Shimizu, H, Toma-Fukai, S, Shimizu, T. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the activation of an Arabidopsis organellar C-to-U RNA editing enzyme by active site complementation.
Plant Cell, 35, 2023
|
|
7BY1
 
 | |
1WY5
 
 | | Crystal structure of isoluecyl-tRNA lysidine synthetase | | Descriptor: | Hypothetical UPF0072 protein AQ_1887 | | Authors: | Nakanishi, K, Fukai, S, Ikeuchi, Y, Soma, A, Sekine, Y, Suzuki, T, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for lysidine formation by ATP pyrophosphatase accompanied by a lysine-specific loop and a tRNA-recognition domain.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1VFG
 
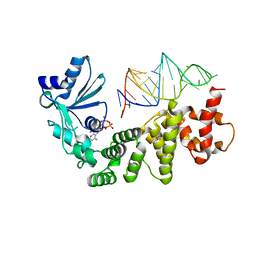 | | Crystal structure of tRNA nucleotidyltransferase complexed with a primer tRNA and an incoming ATP analog | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, RNA (75-MER), poly A polymerase | | Authors: | Tomita, K, Fukai, S, Ishitani, R, Ueda, T, Takeuchi, N, Vassylyev, D.G, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for template-independent RNA polymerization.
Nature, 430, 2004
|
|
5GVC
 
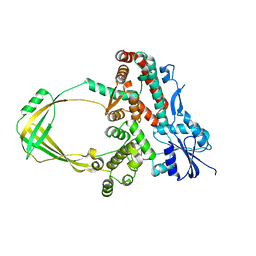 | | Human Topoisomerase IIIb topo domain | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5GVD
 
 | | Human TDRD3 DUF1767-OB domains | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
5GVE
 
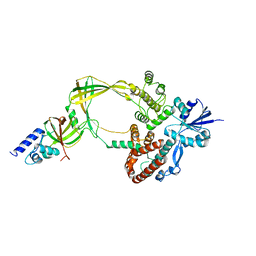 | | Human TOP3B-TDRD3 complex | | Descriptor: | DNA topoisomerase 3-beta-1, MAGNESIUM ION, Tudor domain-containing protein 3 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Takahashi, T.S, Fukai, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.606 Å) | | Cite: | Structural basis of the interaction between Topoisomerase III beta and the TDRD3 auxiliary factor
Sci Rep, 7, 2017
|
|
3WST
 
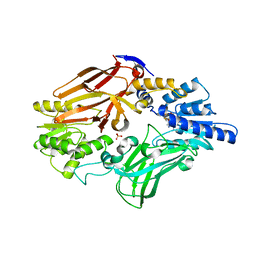 | |
5H11
 
 | |
