8TJF
 
 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | Fab Lambda light chain, IgG1 Fab heavy chain | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y, Chiang, C. | | Deposit date: | 2023-07-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
8TI4
 
 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | GLYCEROL, IgG1 Fab heavy chain, mutated to promote correct pairing, ... | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
6B0F
 
 | |
8WQ4
 
 | |
8WKP
 
 | |
8WQ2
 
 | |
5ZVA
 
 | | APOBEC3F Chimeric Catalytic Domain in Complex with DNA(dC9) | | Descriptor: | APEBEC3F/ssDNA-C9, CACODYLATE ION, DNA (5'-D(*AP*TP*TP*TP*TP*CP*AP*AP*CP*T)-3'), ... | | Authors: | Cheng, C, Zhang, T.L, Wang, C.X, Lan, W.X, Ding, J.P, Cao, C.Y. | | Deposit date: | 2018-05-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Cytidine Deaminase Human APOBEC3F Chimeric Catalytic Domain in Complex with DNA
Chin.J.Chem., 36, 2018
|
|
6LUS
 
 | | Crystal structure of the Mengla Virus VP30 C-terminal domain | | Descriptor: | Minor nucleoprotein VP30 | | Authors: | Dong, S.S, Wen, K.N, Chu, H.G, Wang, C.H, Qin, X.C. | | Deposit date: | 2020-01-30 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Mengla virus VP30 C-terminal domain.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
8HSY
 
 | | Acyl-ACP Synthetase structure | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | An inhibitory mechanism of AasS, an exogenous fatty acid scavenger: Implications for re-sensitization of FAS II antimicrobials.
Plos Pathog., 20, 2024
|
|
8JEU
 
 | |
8JEC
 
 | | plant potassium channel SKOR mutant - L271P/D312N | | Descriptor: | Potassium channel SKOR | | Authors: | Liu, S, Li, S, Tian, C. | | Deposit date: | 2023-05-15 | | Release date: | 2023-08-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure reveals a symmetry reduction of the plant outward-rectifier potassium channel SKOR.
Cell Discov, 9, 2023
|
|
8JET
 
 | |
7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7WGX
 
 | | SARS-CoV-2 spike glycoprotein trimer in closed state after treatment with Cathepsin L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGY
 
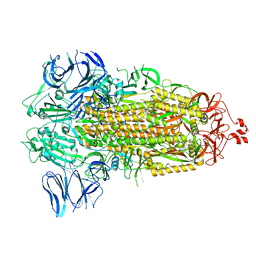 | | SARS-CoV-2 spike glycoprotein trimer in Intermediate state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGZ
 
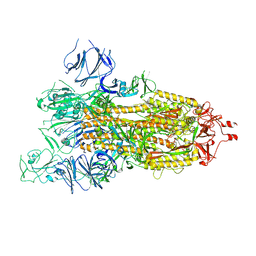 | | SARS-CoV-2 spike glycoprotein trimer in open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7WGV
 
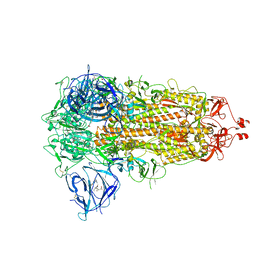 | | SARS-CoV-2 spike glycoprotein trimer in closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Zhu, Y, Tai, L, Yin, G, Sun, F. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Novel cleavage sites identified in SARS-CoV-2 spike protein reveal mechanism for cathepsin L-facilitated viral infection and treatment strategies
Cell Discov, 8, 2022
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
8JNY
 
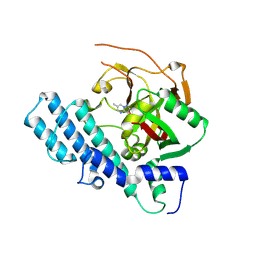 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to a pyrazolopyrimidine carboxamide inhibitor | | Descriptor: | 6-methylpyrazolo[1,5-a]pyrimidine-3-carboxamide, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Wang, C.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Employing a Highly Potent Fluorescence Probe to Discover a PARP-1/2 Binder and the Complex Structures Analysis.
Chemmedchem, 2025
|
|
8JNZ
 
 | | Human ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a pyrazolopyrimidine carboxamide inhibitor | | Descriptor: | 6-methylpyrazolo[1,5-a]pyrimidine-3-carboxamide, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | Authors: | Wang, X.Y, Wang, C.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Employing a Highly Potent Fluorescence Probe to Discover a PARP-1/2 Binder and the Complex Structures Analysis.
Chemmedchem, 2025
|
|
7DLV
 
 | | shrimp dUTPase in complex with Stl | | Descriptor: | CALCIUM ION, Orf20, SULFATE ION, ... | | Authors: | Ma, Q, Wang, F. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of staphylococcal Stl inhibition on a eukaryotic dUTPase.
Int.J.Biol.Macromol., 184, 2021
|
|
7U6R
 
 | |
4HNA
 
 | |
4LNU
 
 | | Nucleotide-free kinesin motor domain in complex with tubulin and a DARPin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Designed ankyrin repeat protein (DARPIN) D1, GLYCEROL, ... | | Authors: | Cao, L, Gigant, B, Knossow, M. | | Deposit date: | 2013-07-12 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement
Nat Commun, 5, 2014
|
|
8WWC
 
 | | De novo design binder of HRAS -120-4 | | Descriptor: | De novo design protein 120-4, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Wang, L, Wang, S, Liu, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
