2G03
 
 | | Structure of a putative cell filamentation protein from Neisseria meningitidis. | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, hypothetical protein NMA0004 | | Authors: | Cuff, M.E, Bigelow, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative cell filamentation protein from Neisseria meningitidis.
To be Published
|
|
2FU6
 
 | |
6DW3
 
 | | SAMHD1 Bound to Cytarabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 4-amino-1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DXP
 
 | | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Arcinas, A.J, Ghosh, A, Chamala, S, Bonanno, J.B, Kelly, L, Almo, S.C. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae
To Be Published
|
|
2FXZ
 
 | |
6DUC
 
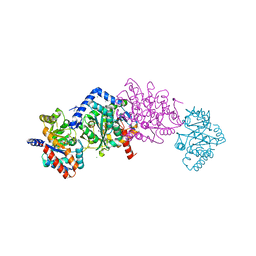 | | Crystal structure of mutant beta-K167T tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, cesium ion at the metal coordination site, and 2-aminophenol quinonoid (1D0) at the beta-site | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-3-[(2-hydroxyphenyl)amino]propanoic acid, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Tryptophan synthase Q114A mutant in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate (P1T) at the beta site, and cesium ion at the metal coordination site.
To be Published
|
|
6WJ5
 
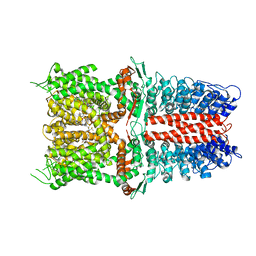 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
4LS3
 
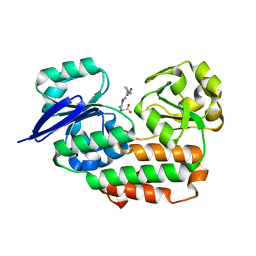 | | THE crystal STRUCTURE OF HELICOBACTER PYLORI CEUE(HP1561)/NI-HIS COMPL | | Descriptor: | HISTIDINE, NICKEL (II) ION, Nickel (III) ABC transporter, ... | | Authors: | Salamina, M, Shaik, M.M, Cendron, L, Zanotti, G. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Mol.Microbiol., 91, 2014
|
|
9FJN
 
 | | Solution NMR structure of a peptide encompassing residues 2-19 of the human formin INF2 | | Descriptor: | Inverted formin-2 | | Authors: | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
4M64
 
 | |
4M7D
 
 | | Crystal structure of Lsm2-8 complex bound to the RNA fragment CGUUU | | Descriptor: | U6 snRNA, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4LY6
 
 | | Nucleotide-induced asymmetry within ATPase activator ring drives s54-RNAP interaction and ATP hydrolysis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family), ... | | Authors: | Sysoeva, T.A, Chowdhury, S, Guo, L, Nixon, B.T. | | Deposit date: | 2013-07-30 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Nucleotide-induced asymmetry within ATPase activator ring drives sigma 54-RNAP interaction and ATP hydrolysis.
Genes Dev., 27, 2013
|
|
4MBR
 
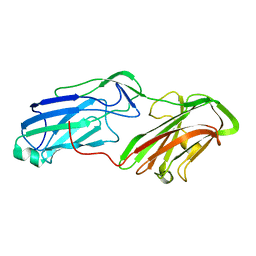 | | 3.65 Angstrom Crystal Structure of Serine-rich Repeat Protein (Srr2) from Streptococcus agalactiae | | Descriptor: | Serine-rich repeat protein 2 | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Seo, H.S, Seepersaud, R, Doran, K.S, Iverson, T.M, Sullam, P.M, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Characterization of Fibrinogen Binding by Glycoproteins Srr1 and Srr2 of Streptococcus agalactiae.
J.Biol.Chem., 288, 2013
|
|
4M6W
 
 | | Crystal structure of the C-terminal segment of FANCM in complex with FAAP24 | | Descriptor: | Fanconi anemia group M protein, Fanconi anemia-associated protein of 24 kDa, SULFATE ION | | Authors: | Yang, H, Zhang, T, Tong, L, Ding, J. | | Deposit date: | 2013-08-11 | | Release date: | 2013-10-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair.
Nucleic Acids Res., 41, 2013
|
|
4M75
 
 | | Crystal structure of Lsm1-7 complex | | Descriptor: | CHLORIDE ION, U6 snRNA-associated Sm-like protein Lsm1, U6 snRNA-associated Sm-like protein Lsm2, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4LP8
 
 | | A Novel Open-State Crystal Structure of the Prokaryotic Inward Rectifier KirBac3.1 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Inward rectifier potassium channel Kirbac3.1, ... | | Authors: | Zubcevic, L, Bavro, V.N, Muniz, J.R.C, Schmidt, M.R, Wang, S, De Zorzi, R, Venien-Bryan, C, Sansom, M.S.P, Nichols, C.G, Tucker, S.J. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Control of KirBac3.1 Potassium Channel Gating at the Interface between Cytoplasmic Domains.
J.Biol.Chem., 289, 2014
|
|
9FJW
 
 | | Solution NMR structure of a peptide encompassing residues 2-36 of the human formin INF2 | | Descriptor: | Inverted formin-2 | | Authors: | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
4LS0
 
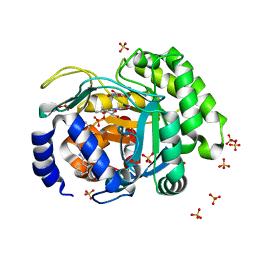 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH01B0033 | | Descriptor: | 2-{(E)-[2-(4-phenyl-1,3-thiazol-2-yl)hydrazinylidene]methyl}benzaldehyde, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DHO1B0033
To be Published
|
|
4LT5
 
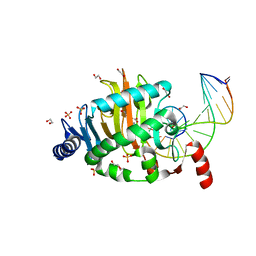 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | Authors: | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
4LP4
 
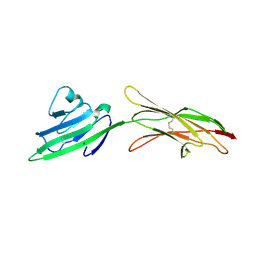 | |
4LVD
 
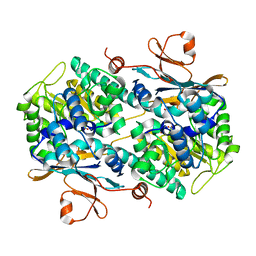 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-nitrophenyl)cyclopropanecarboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4M02
 
 | | Middle fragment(residues 494-663) of the binding region of SraP | | Descriptor: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4M77
 
 | | Crystal structure of Lsm2-8 complex, space group I212121 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3, U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Zhou, L, Hang, J, Zhou, Y, Wan, R, Lu, G, Yan, C, Shi, Y. | | Deposit date: | 2013-08-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Crystal structures of the Lsm complex bound to the 3' end sequence of U6 small nuclear RNA.
Nature, 506, 2014
|
|
4MAD
 
 | | Crystal structure of beta-galactosidase C (BgaC) from Bacillus circulans ATCC 31382 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-galactosidase | | Authors: | Kamerke, C, You, D.J, Kanaya, S, Elling, L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of a glycosynthase by the crystal structure of beta-galactosidase from Bacillus circulans (BgaC) and its use for the synthesis of N-acetyllactosamine type 1 glycan structures.
J.Biotechnol., 191, 2014
|
|
4MCU
 
 | |
