8HFI
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant desipramine in an inward-open state at resolution of 2.5 angstrom. | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8HFL
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant bupropion in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | Bupropion, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8HFG
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of dopamine in an inward-open state at resolution of 3.0 angstrom. | | Descriptor: | CHLORIDE ION, L-DOPAMINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8HFE
 
 | | Cryo-EM structure of human norepinephrine transporter NET in an inward-open state at resolution of 2.5 angstrom | | Descriptor: | CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
4YJE
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A1 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
4YJL
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A2 | | Descriptor: | 1,2-ETHANEDIOL, APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
4YK6
 
 | | Crystal structure of APC-ARM in complexed with Amer1-A4 | | Descriptor: | APC membrane recruitment protein 1, Adenomatous polyposis coli protein | | Authors: | Zhang, Z, Xiao, Y, Wu, G. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the APC-ARM domain in complexes with discrete Amer1/WTX fragments reveal that it uses a consensus mode to recognize its binding partners
Cell Discov, 1, 2015
|
|
5HQB
 
 | | A Glycoside Hydrolase Family 97 enzyme (E480Q) in complex with Panose from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HQA
 
 | | A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, CALCIUM ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
4ZM6
 
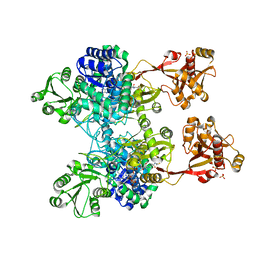 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
5HQ4
 
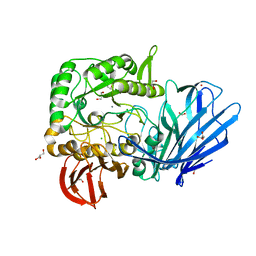 | | A Glycoside Hydrolase Family 97 enzyme from Pseudoalteromonas sp. strain K8 | | Descriptor: | Alpha-glucosidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5HQC
 
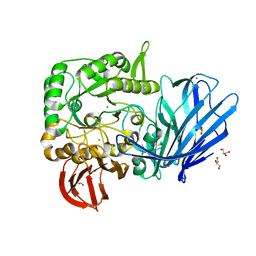 | | A Glycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5H9F
 
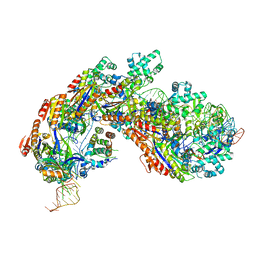 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target at 2.45 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
1U9S
 
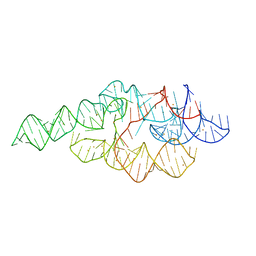 | | Crystal structure of the specificity domain of Ribonuclease P of the A-type | | Descriptor: | BARIUM ION, RIBONUCLEASE P | | Authors: | Krasilnikov, A.S, Xiao, Y, Pan, T, Mondragon, A. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Basis for structural diversity in homologous RNAs.
Science, 306, 2004
|
|
3VCP
 
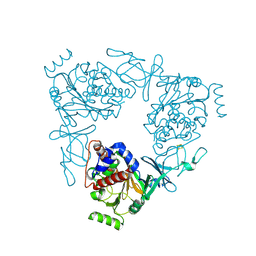 | | The 2.2 Angstrom structure of Stc2 with proline bound in the active site | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, PROLINE, ... | | Authors: | Daughtry, K.D, Xiao, Y, Stoner-Ma, D, Cho, E, Orville, A.M, Liu, P, Allen, K.N. | | Deposit date: | 2012-01-04 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman, and UV-Visible Spectroscopic Analysis of a Rieske-Type Demethylase.
J.Am.Chem.Soc., 134, 2012
|
|
3O5Z
 
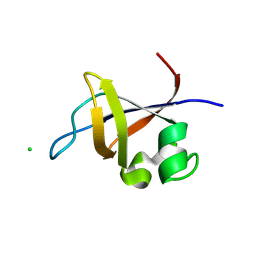 | | Crystal structure of the SH3 domain from p85beta subunit of phosphoinositide 3-kinase (PI3K) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Chen, S, Xiao, Y, Ponnusamy, R, Tan, J, Lei, J, Hilgenfeld, R. | | Deposit date: | 2010-07-28 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-ray structure of the SH3 domain of the phosphoinositide 3-kinase p85 beta subunit
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5H9E
 
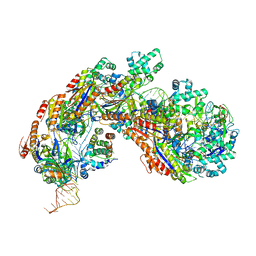 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target (32-nt spacer) at 3.20 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
3VCA
 
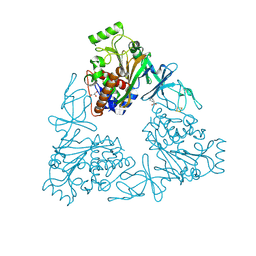 | | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman and UV-visible Spectroscopic Analysis of a Rieske-type Demethylase | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Daughtry, K.D, Xiao, Y, Stoner-Ma, D, Cho, E, Orville, A.M, Liu, P, Allen, K.N. | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman, and UV-Visible Spectroscopic Analysis of a Rieske-Type Demethylase.
J.Am.Chem.Soc., 134, 2012
|
|
4F7K
 
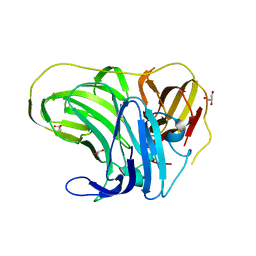 | |
2YNA
 
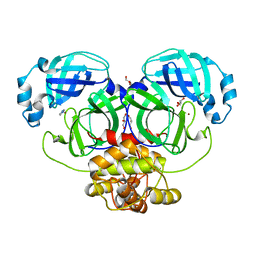 | |
2YNB
 
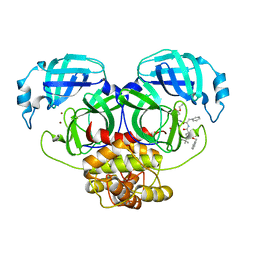 | |
4JNM
 
 | | Discovery of Potent and Efficacious Urea-containing Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors with Reduced CYP2C9 Inhibition Properties | | Descriptor: | 1,2-ETHANEDIOL, 1-[(6-aminopyridin-3-yl)methyl]-3-[4-(phenylsulfonyl)phenyl]urea, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Gunzner-Toste, J, Zhao, G, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Fu, B, Han, B, Ho, Y, Kley, N, Liederer, B, Lin, J, Mukadam, S, O'Brien, T, Reynolds, D.J, Sharma, G, Skelton, N, Smith, C.C, Oh, A, Wang, W, Wang, Z, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X, Bair, K.W, Dragovich, P.S. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of potent and efficacious urea-containing nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with reduced CYP2C9 inhibition properties.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3SJM
 
 | | Crystal Structure Analysis of TRF2-Dbd-DNA complex | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*GP*A)-3'), Telomeric repeat-binding factor 2 | | Authors: | Nair, S.K, Sliverman, S.K, Chen, J.H, Xiao, Y. | | Deposit date: | 2011-06-21 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure Analysis of TRF2-Dbd-DNA complex
To be Published
|
|
4LVF
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-2-phenyl-N-(pyridin-4-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LVA
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[4-(pyrrolidin-1-yl)piperidin-1-yl]sulfonyl}benzyl)-2H-pyrido[4,3-e][1,2,4]thiadiazin-3-amine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
