8Y1J
 
 | |
9EOW
 
 | |
2GA2
 
 | | h-MetAP2 complexed with A193400 | | Descriptor: | 5-BROMO-2-{[(4-CHLOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of sulfonamide compounds as potent methionine aminopeptidase type II inhibitors with antiproliferative properties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8DMD
 
 | |
8EUA
 
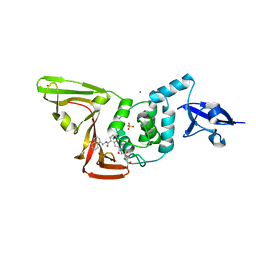 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | Descriptor: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | Authors: | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | Deposit date: | 2022-10-18 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YVR
 
 | | Crystal Structure of L-Threonine Aldolase from Neptunomonas marina | | Descriptor: | GLYCEROL, L-threonine aldolase | | Authors: | He, Y.Z, Wang, J, Yan, W.P, Zhang, Y, Feng, Y. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Engineering of the L-Threonine Aldolase from Neptunomonas marine for the Efficient Synthesis of beta-Hydroxy-alpha-amino Acids via C-C Formation
Acs Catalysis, 2023
|
|
6VLT
 
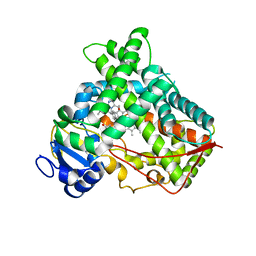 | | Crystal Structure of Human P450 2C9*2 Genetic Variant in Complex with Losartan | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2C9, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shah, M.B. | | Deposit date: | 2020-01-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure of Cytochrome P450 2C9*2 in Complex with Losartan: Insights into the Effect of Genetic Polymorphism.
Mol.Pharmacol., 98, 2020
|
|
8IQ0
 
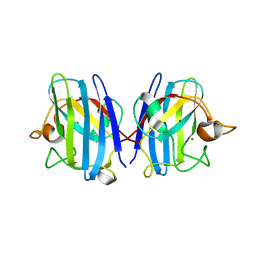 | | Crystal structure of hydrogen sulfide-bound superoxide dismutase in oxidized state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Zhou, J.H, Huang, W.X, Cheng, R.X, Zhang, P.J, Zhu, Y.C. | | Deposit date: | 2023-03-15 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrogen sulfide functions as a micro-modulator bound at the copper active site of Cu/Zn-SOD to regulate the catalytic activity of the enzyme.
Cell Rep, 42, 2023
|
|
8IQ1
 
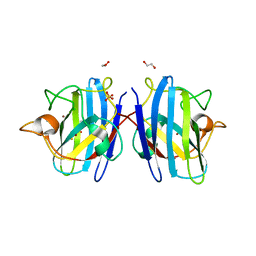 | | Crystal structure of hydrogen sulfide-bound superoxide dismutase in reduced state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Zhou, J.H, Huang, W.X, Cheng, R.X, Zhang, P.J, Zhu, Y.C. | | Deposit date: | 2023-03-15 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen sulfide functions as a micro-modulator bound at the copper active site of Cu/Zn-SOD to regulate the catalytic activity of the enzyme.
Cell Rep, 42, 2023
|
|
8JWY
 
 | | Crystal structure of A2AR-T4L in complex with 2-118 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[2-oxidanylidene-1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]pyridin-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
8JWZ
 
 | | Crystal structure of A2AR-T4L in complex with AB928 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
3KVD
 
 | | Crystal structure of the Neisseria meningitidis Factor H binding protein, fHbp (GNA1870) at 2.0 A resolution | | Descriptor: | Lipoprotein | | Authors: | Cendron, L, Veggi, D, Girardi, E, Zanotti, G. | | Deposit date: | 2009-11-30 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the uncomplexed Neisseria meningitidis factor H-binding protein fHbp (rLP2086).
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5UFG
 
 | | Crystal Structure of CYP2B6 (Y226H/K262R/I114V) in complex with myrtenyl bromide | | Descriptor: | (1S,5R)-2-(bromomethyl)-6,6-dimethylbicyclo[3.1.1]hept-2-ene, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Halpert, J.R. | | Deposit date: | 2017-01-04 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Halogen-pi Interactions in the Cytochrome P450 Active Site: Structural Insights into Human CYP2B6 Substrate Selectivity.
ACS Chem. Biol., 12, 2017
|
|
5UEC
 
 | | Crystal Structure of CYP2B6 (Y226H/K262R) in complex with myrtenyl bromide. | | Descriptor: | (1S,5R)-2-(bromomethyl)-6,6-dimethylbicyclo[3.1.1]hept-2-ene, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Halpert, J.R. | | Deposit date: | 2016-12-30 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Halogen-pi Interactions in the Cytochrome P450 Active Site: Structural Insights into Human CYP2B6 Substrate Selectivity.
ACS Chem. Biol., 12, 2017
|
|
5UDA
 
 | |
5TPG
 
 | | Optimization of spirocyclic proline tryptophanhydroxylase-1 inhibitors | | Descriptor: | (3S)-8-(2-amino-6-{(1R)-1-[5-chloro-3'-(methylsulfonyl)[1,1'-biphenyl]-2-yl]-2,2,2-trifluoroethoxy}pyrimidin-4-yl)-2,8-diazaspiro[4.5]decane-3-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETONITRILE, ... | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UNI
 
 | |
5T0O
 
 | | Crystal Structure of a membrane protein | | Descriptor: | CmeB | | Authors: | Su, C.-C, Yu, E.W. | | Deposit date: | 2016-08-16 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump.
Nat Commun, 8, 2017
|
|
5UAP
 
 | | Crystal Structure of CYP2B6 (Y226H/K262R) in complex with Bornyl Bromide | | Descriptor: | (1R,2R,4R)-2-bromo-1,7,7-trimethylbicyclo[2.2.1]heptane, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Halpert, J.R. | | Deposit date: | 2016-12-19 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Halogen-pi Interactions in the Cytochrome P450 Active Site: Structural Insights into Human CYP2B6 Substrate Selectivity.
ACS Chem. Biol., 12, 2017
|
|
5ZO0
 
 | | Neutron structure of xylanase at pD5.4 | | Descriptor: | Endo-1,4-beta-xylanase 2 | | Authors: | Wan, Q, Li, Z.H. | | Deposit date: | 2018-04-12 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.648 Å) | | Cite: | Neutron structure of xylanase at pD5.4
To be published
|
|
7PT0
 
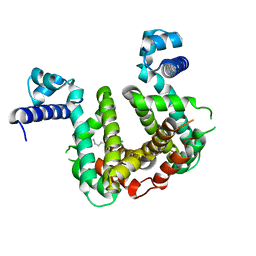 | | SCO3201 with putative ligand | | Descriptor: | SPERMIDINE, TetR family transcriptional regulator | | Authors: | Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2021-09-25 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of free and ligand-bound forms of the TetR/AcrR-like regulator SCO3201 from Streptomyces coelicolor suggest a novel allosteric mechanism.
Febs J., 290, 2023
|
|
6RIC
 
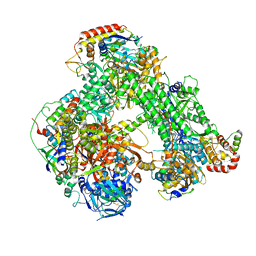 | | Structure of the core Vaccinia Virus DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, H.S, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
6K9R
 
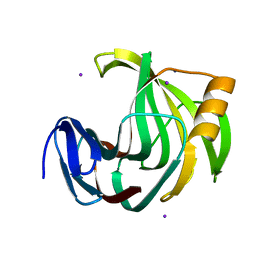 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
8HUG
 
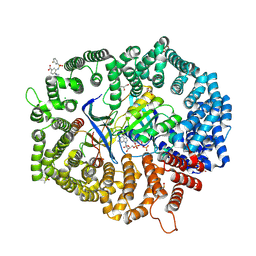 | | F1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 4-[4-(3-chlorophenyl)piperazin-1-yl]-3-[(3-fluorophenyl)sulfonylamino]benzoic acid, CHLORIDE ION, CRM1 isoform 1, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HUF
 
 | | B28 in complex with CRM1-Ran-RanBP1 | | Descriptor: | 3-[(4-chlorophenyl)carbonylamino]-4-[4-(2,5-dimethylphenyl)piperazin-1-yl]benzoic acid, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of a Hidden Pocket beneath the NES Groove by Novel Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
