8K0H
 
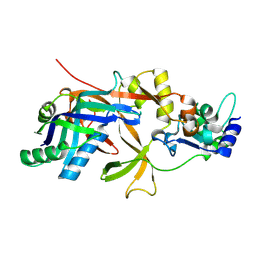 | | Structure of Cas5f-8f | | Descriptor: | Csy1, Csy2 | | Authors: | Feng, Y, Wang, H. | | Deposit date: | 2023-07-09 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Cas1 mediates the interference stage in a phage-encoded CRISPR-Cas system.
Nat.Chem.Biol., 20, 2024
|
|
8K0J
 
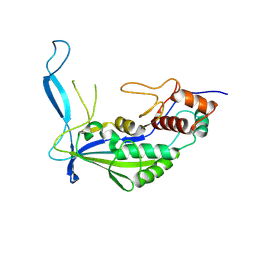 | | Crystal structure of Cas7f | | Descriptor: | Csy3 | | Authors: | Feng, Y, Wang, H. | | Deposit date: | 2023-07-09 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.365 Å) | | Cite: | Cas1 mediates the interference stage in a phage-encoded CRISPR-Cas system.
Nat.Chem.Biol., 20, 2024
|
|
8K0K
 
 | | Crystal structure of Csy complex | | Descriptor: | Csy1, Csy2, Csy3, ... | | Authors: | Feng, Y, Wang, H. | | Deposit date: | 2023-07-09 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Cas1 mediates the interference stage in a phage-encoded CRISPR-Cas system.
Nat.Chem.Biol., 20, 2024
|
|
7WC5
 
 | | Crystal structure of serotonin 2A receptor in complex with psilocin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-(dimethylamino)ethyl]-1~{H}-indol-4-ol, 5-hydroxytryptamine receptor 2A, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC6
 
 | | Crystal structure of serotonin 2A receptor in complex with LSD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC9
 
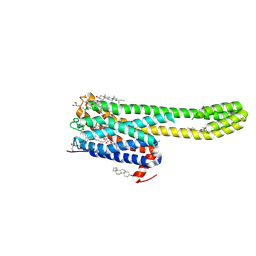 | | Crystal structure of serotonin 2A receptor in complex with non-hallucinogenic psychedelic analog | | Descriptor: | (10~{R},15~{S})-12-[3-(2-methoxyphenyl)propyl]-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5(16),6,8-triene, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC7
 
 | | Crystal structure of serotonin 2A receptor in complex with lisuride | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
7WC8
 
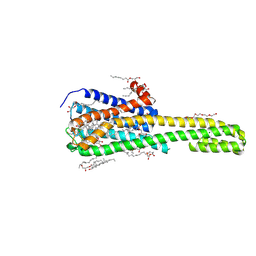 | | Crystal structure of serotonin 2A receptor in complex with lumateperone | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(10~{R},15~{S})-4-methyl-1,4,12-triazatetracyclo[7.6.1.0^{5,16}.0^{10,15}]hexadeca-5,7,9(16)-trien-12-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | Deposit date: | 2021-12-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
4KAJ
 
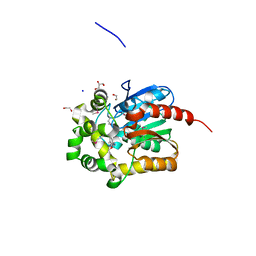 | | X-Ray Structure of the complex of Haloalkane dehalogenase HaloTag7 with HALTS, Northeast Structural Genomics Consortium (NESG) Target OR151 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kuzin, A, Lew, S, Neklesa, T.K, Noblin, D, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Crews, C.M, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-22 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR151
To be Published
|
|
4KAC
 
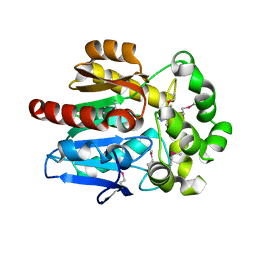 | | X-Ray Structure of the complex HaloTag2 with HALTS. Northeast Structural Genomics Consortium (NESG) Target OR150. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AMMONIUM ION, Haloalkane dehalogenase, ... | | Authors: | Kuzin, A, Lew, S, Neklesa, T.K, Noblin, D, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Crews, C.M, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-22 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR150
To be Published
|
|
9KQC
 
 | | Chlorella virus Hyaluronan Synthase bound to VNAR | | Descriptor: | Hyaluronan synthase, MANGANESE (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, ... | | Authors: | Deng, P, Zhang, X, Xu, M, Wen, J, Li, P, Bi, Y, Wang, H. | | Deposit date: | 2024-11-25 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Generation of shark single-domain antibodies as an aid for Cryo-EM structure determination of membrane proteins: Use hyaluronan synthase as an example.
J Struct Biol X, 11, 2025
|
|
4KYV
 
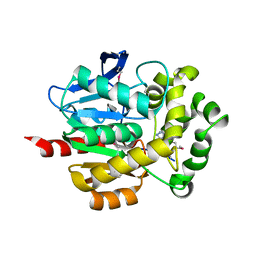 | | Crystal Structure of dehalogenase HaloTag2 with HALTS at the resolution 1.8A. Northeast Structural Genomics Consortium (NESG) Target OR150 | | Descriptor: | N-(2-ethoxy-3,5-dimethylbenzyl)-1H-tetrazol-5-amine, SODIUM ION, dehalogenase HaloTag2 | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR150
To be Published
|
|
4L6E
 
 | | Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2. Northeast Structural Genomics Consortium (NESG) Target HR9193b | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, L, Xiao, R, Maglaqui, M, Kogan, S, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-12 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Crystal Structure of the RanBD1 fourth domain of E3 SUMO-protein ligase RanBP2.
To be Published
|
|
2LG2
 
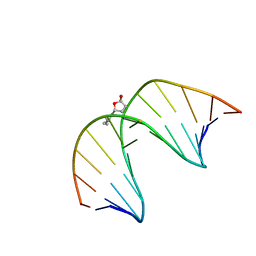 | | Structure of the duplex containing HNE derived (6S,8R,11S) N2-dG cyclic hemiacetal when placed opposite dT | | Descriptor: | (2R,5S)-5-pentyltetrahydrofuran-2-ol, DNA (5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*GP*CP*TP*AP*GP*C)-3') | | Authors: | Huang, H, Wang, H, Kozekova, A, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ring-chain tautomerization of trans-4-hydroxynonenal derived (6S,8R,11S) gamma-hydroxy-1,N2-propano-deoxyguanosine adduct when placed opposite 2'-deoxythymidine in duplex
To be Published
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7CXM
 
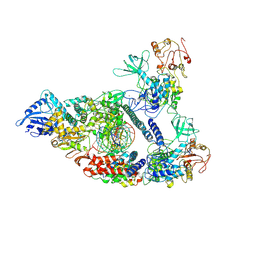 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7HSC
 
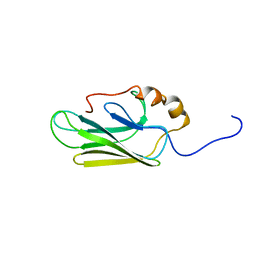 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE HEAT SHOCK COGNATE-70 KD SUBSTRATE BINDING DOMAIN OBTAINED BY MULTIDIMENSIONAL NMR TECHNIQUES | | Descriptor: | PROTEIN (HEAT SHOCK COGNATE 70 KD PROTEIN 1) | | Authors: | Morshauser, R.C, Hu, W, Wang, H, Pang, Y, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70.
J.Mol.Biol., 289, 1999
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
8WDM
 
 | | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis | | Descriptor: | Carboxylic ester hydrolase | | Authors: | Li, Z.S, Wang, H, Gao, J, Chen, Y.Y, Wei, H.L, Li, Q, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis
To Be Published
|
|
5X9J
 
 | |
3TKT
 
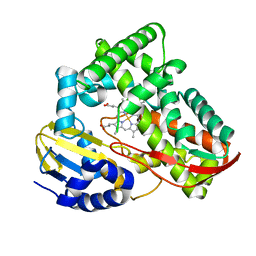 | | Crystal structure of CYP108D1 from Novosphingobium aromaticivorans DSM12444 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2011-08-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of CYP108D1 from Novosphingobium aromaticivorans DSM12444: an aromatic hydrocarbon-binding P450 enzyme
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2L7K
 
 | | Solution NMR Structure of protein CD1104.2 from Clostridium difficile, Northeast Structural Genomics Consortium Target CfR130 | | Descriptor: | Uncharacterized protein | | Authors: | Pulavarti, S.V.S.R.K, Eletsky, A, Mills, J.L, Sukumaran, D.K, Wang, H, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-13 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CfR130 from Clostridium difficile, Northeast Structural Genomics Consortium Target CfR130
To be Published
|
|
7YPN
 
 | | Crystal structure of transaminase CC1012 mutant M9 complexed with PLP | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, L, Wang, H, Wei, D. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism-Guided Computational Design of omega-Transaminase by Reprograming of High-Energy-Barrier Steps.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3SZ5
 
 | | Crystal Structure of LHK-Exo in complex with 5-phosphorylated oligothymidine (dT)4 | | Descriptor: | 5'-D(P*TP*TP*TP*T)-3', Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3SYY
 
 | | Crystal Structure of an alkaline exonuclease (LHK-Exo) from Laribacter hongkongensis | | Descriptor: | Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
