4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
3SHZ
 
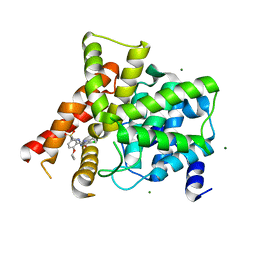 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-chloro-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SHY
 
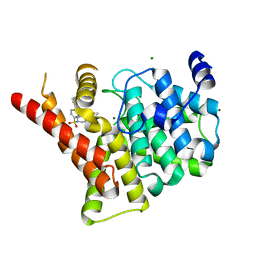 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-fluoro-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SIE
 
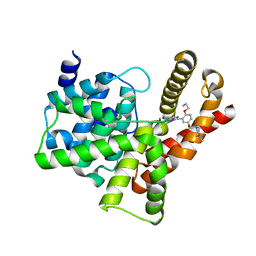 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
5UMY
 
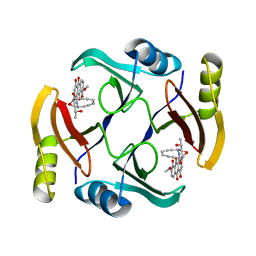 | | Crystal structure of TnmS3 in complex with tiancimycin | | Descriptor: | (1aS,11S,11aR,14Z,18R)-3,8,18-trihydroxy-11a-[(1R)-1-hydroxyethyl]-7-methoxy-11,11a-dihydro-4H-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinoline-4,9(10H)-dione, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
8HE4
 
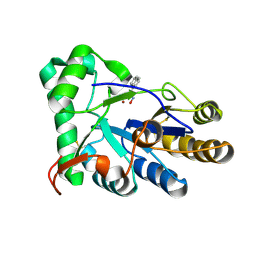 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE2
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE1
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
 | |
8HF9
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
5VC8
 
 | | Crystal structure of the WHSC1 PWWP1 domain | | Descriptor: | DNA (5'-D(P*CP*TP*(DN))-3'), Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
4UEI
 
 | | Solution structure of the sterol carrier protein domain 2 of Helicoverpa armigera | | Descriptor: | STEROL CARRIER PROTEIN 2/3-OXOACYL-COA THIOLASE | | Authors: | Liu, X, Ma, H, Yan, X, Hong, H, Peng, J, Peng, R. | | Deposit date: | 2014-12-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Function of Helicoverpa Armigera Sterol Carrier Protein-2, an Important Insecticidal Target from the Cotton Bollworm.
Sci.Rep., 5, 2015
|
|
5UMQ
 
 | | Crystal structure of TnmS1, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMX
 
 | | Crystal structure of TnmS3 in complex with riboflavin | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMP
 
 | | Crystal structure of TnmS3, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMW
 
 | | Crystal structure of TnmS2, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
8SIV
 
 | | Structure of Compound 2 bound to the CHK1 10-point mutant | | Descriptor: | N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)cyclopropanecarboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8SIX
 
 | | Structure of Compound 13 bound to the CHK1 10-point mutant | | Descriptor: | (1S)-N-(7-chloro-6-{4-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperazin-1-yl}isoquinolin-3-yl)-6-oxaspiro[2.5]octane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8SIW
 
 | | Structure of Compound 5 bound to the CHK1 10-point mutant | | Descriptor: | (1S,2S)-N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)-2-(1-methyl-1H-pyrazol-4-yl)cyclopropane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8E80
 
 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 14 | | Descriptor: | 2-[(1r,4r)-2-{(6P)-6-[(6M)-6-(1H-pyrazol-5-yl)-1H-indazol-1-yl]pyrimidin-4-yl}-2-azabicyclo[2.1.1]hexan-4-yl]propan-2-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
8E81
 
 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 25 | | Descriptor: | (1S)-1-[(1P)-1-{6-[(3R)-3-(2-hydroxypropan-2-yl)pyrrolidin-1-yl]pyrimidin-4-yl}-1H-indazol-6-yl]spiro[2.2]pentane-1-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
8JRU
 
 | | Cryo-EM structure of the glucagon receptor bound to beta-arrestin 1 in ligand-free state | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, Nanobody 32, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
8JRV
 
 | | Cryo-EM structure of the glucagon receptor bound to glucagon and beta-arrestin 1 | | Descriptor: | Beta-arrestin 1 and single-chain fragment variable 30 (scFv30), Glucagon, HA signal peptide,HPC4 purification tag,Glucagon receptor,C-terminal tail of Vasopressin V2 receptor, ... | | Authors: | Chen, K, Zhang, C, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tail engagement of arrestin at the glucagon receptor.
Nature, 620, 2023
|
|
8JVA
 
 | | Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 heavy chain, S2L20 light chain, ... | | Authors: | Liu, B, Liu, H.H, Han, P, Qi, J.X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Enhanced potency of an IgM-like nanobody targeting conserved epitope in SARS-CoV-2 spike N-terminal domain.
Signal Transduct Target Ther, 9, 2024
|
|
7CRZ
 
 | | Crystal structure of human glucose transporter GLUT3 bound with C3361 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R,4S,5R,6R)-6-(hydroxymethyl)-4-undec-10-enoxy-oxane-2,3,5-triol, Solute carrier family 2, ... | | Authors: | Yuan, Y.Y, Zhang, S, Wang, N, Jiang, X, Yan, N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orthosteric-allosteric dual inhibitors of PfHT1 as selective antimalarial agents.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
