2BE0
 
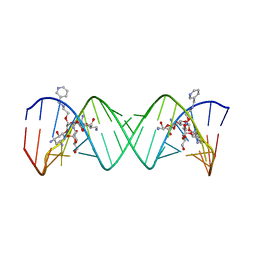 | | Complex Between Paromomycin Derivative JS5-39 and the 16S-Rrna A-Site. | | Descriptor: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R,4R,5S,6R)-3-AMINO-4 ,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-(HYDROXYMETHYL)-4-(2-((R)-PIPERIDI N-3-YLMETHYLAMINO)ETHOXY)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Francois, B, Westhof, E. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
3JUJ
 
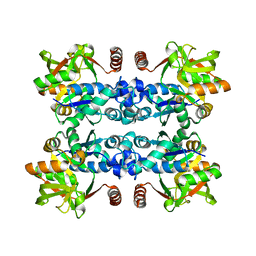 | |
7Z6R
 
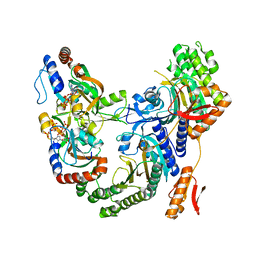 | | Psychrobacter arcticus ATPPRT (HisGZ) R56A mutant bound to ATP and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Fisher, G, da Silva, R.G. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric rescue of catalytically impaired ATP phosphoribosyltransferase variants links protein dynamics to active-site electrostatic preorganisation.
Nat Commun, 13, 2022
|
|
7Z8U
 
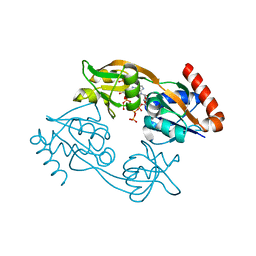 | | Catalytic subunit HisG R56A mutant from Psychrobacter arcticus ATPPRT (HisGZ) in complex with ATP and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Fisher, G, da Silva, R.G. | | Deposit date: | 2022-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric rescue of catalytically impaired ATP phosphoribosyltransferase variants links protein dynamics to active-site electrostatic preorganisation.
Nat Commun, 13, 2022
|
|
3JUK
 
 | |
1OM1
 
 | | Crystal structure of maize CK2 alpha in complex with IQA | | Descriptor: | (5-OXO-5,6-DIHYDRO-INDOLO[1,2-A]QUINAZOLIN-7-YL)-ACETIC ACID, Casein kinase II, alpha chain | | Authors: | Battistutta, R, De Moliner, E, Zanotti, G. | | Deposit date: | 2003-02-24 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Biochemical and three-dimensional-structural study of the specific inhibition of protein kinase CK2 by [5-oxo-5,6-dihydroindolo-(1,2-a)quinazolin-7-yl]acetic acid (IQA).
Biochem.J., 374, 2003
|
|
7Z6B
 
 | |
7LGS
 
 | |
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
7KEF
 
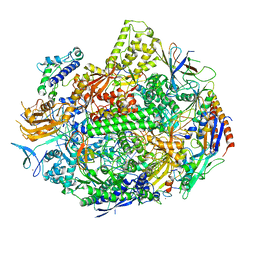 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaM in swing state | | Descriptor: | (1S)-1,4-anhydro-1-(3-methoxynaphthalen-2-yl)-5-O-phosphono-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KEE
 
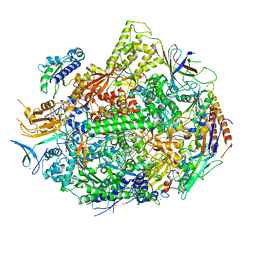 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaMTP bound to E-site | | Descriptor: | (1S)-1,4-anhydro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-1-(3-methoxynaphthalen-2-yl)-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KED
 
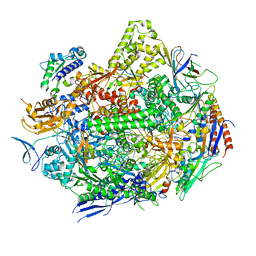 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, W, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
4JYP
 
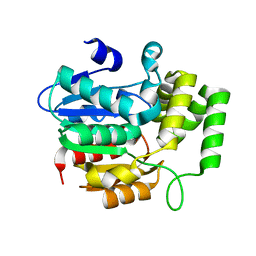 | | crystal Structure of KAI2 Apo form | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Guo, Y, Zheng, Z, Noel, J.P. | | Deposit date: | 2013-03-31 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYM
 
 | | crystal Structure of KAI2 in complex with 3-methyl-2H-furo[2,3-c]pyran-2-one | | Descriptor: | 3-methyl-2H-furo[2,3-c]pyran-2-one, Hydrolase, alpha/beta fold family protein | | Authors: | Guo, Y, Zheng, Z, Noel, J.P. | | Deposit date: | 2013-03-30 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3QSD
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with CA074 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
1PJV
 
 | | Cobatoxin 1 from Centruroides noxius Scorpion venom: Chemical Synthesis, 3-D Structure in Solution, Pharmacology and Docking on K+ channels | | Descriptor: | Cobatoxin 1 | | Authors: | Mosbah, A, Jouirou, B, Visan, V, Grissmer, S, El Ayeb, M, Rochat, H, De Waard, M, Mabrouk, K, Sabatier, J.M. | | Deposit date: | 2003-06-03 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Cobatoxin 1 from Centruroides noxius scorpion venom: chemical synthesis, three-dimensional structure in solution, pharmacology and docking on K+ channels.
Biochem.J., 377, 2004
|
|
3S3Q
 
 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with K11017 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-05-18 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
7K7A
 
 | | Transmembrane structure of TNFR1 | | Descriptor: | Tumor necrosis factor receptor superfamily member 1A | | Authors: | Zhao, L, Chou, J. | | Deposit date: | 2020-09-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Diversity and Similarity of Transmembrane Trimerization of TNF Receptors.
Front Cell Dev Biol, 8, 2020
|
|
3S3R
 
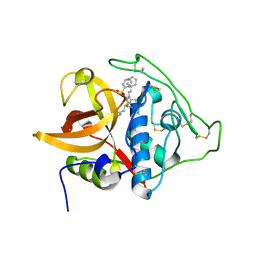 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with K11777 inhibitor | | Descriptor: | Cathepsin B-like peptidase (C01 family), Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-05-18 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
4GR6
 
 | | Crystal structure of AtRbcX2 from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, AtRbcX2 | | Authors: | Grudnik, P, Golik, P, Kolesinski, P, Dubin, G, Szczepaniak, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into eukaryotic Rubisco assembly - Crystal structures of RbcX chaperones from Arabidopsis thaliana.
Biochim.Biophys.Acta, 1830, 2013
|
|
4GR2
 
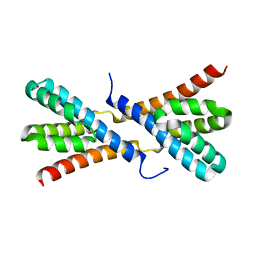 | | Structure of AtRbcX1 from Arabidopsis thaliana. | | Descriptor: | AtRbcX1 | | Authors: | Golik, P, Grudnik, P, Kolesinski, P, Dubin, G, Szczepaniak, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into eukaryotic Rubisco assembly - Crystal structures of RbcX chaperones from Arabidopsis thaliana.
Biochim.Biophys.Acta, 1830, 2013
|
|
1RUO
 
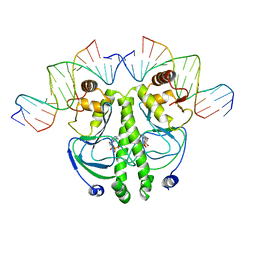 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP) MUTANT/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
8SY5
 
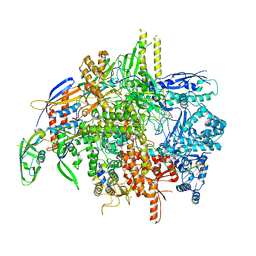 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dS-BTP base pair in the active site | | Descriptor: | 2-oxo-2-hydroadenosine 5'-(tetrahydrogen triphosphate), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY6
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-UTP base pair in the active site | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY7
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-STP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
