8SUR
 
 | | TMEM16F bound with Niclosamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide, ... | | Authors: | Feng, S, Cheng, Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8TAG
 
 | | TMEM16F, with Calcium and PIP2, no inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Cheng, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
3V35
 
 | | Aldose reductase complexed with a nitro compound | | Descriptor: | 2-[(5-nitro-1,3-thiazol-2-yl)carbamoyl]phenyl acetate, Aldose reductase, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | Deposit date: | 2011-12-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
8SUN
 
 | | TMEM16F 1PBC | | Descriptor: | 1-Hydroxy-3-(trifluoromethyl)pyrido[1,2-a]benzimidazole-4-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, ... | | Authors: | Wu, H, Feng, S, Cheng, Y. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8JUN
 
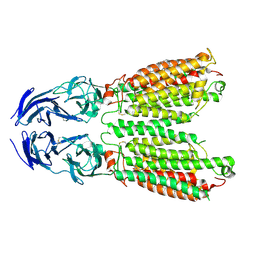 | | Cryo-EM structure of SIDT1 E555Q mutant | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SID1 transmembrane family member 1, ZINC ION | | Authors: | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
8JUL
 
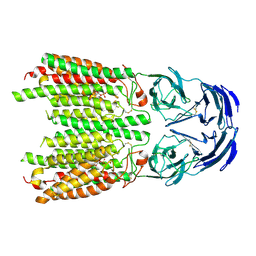 | | Cryo-EM structure of SIDT1 in complex with phosphatidic acid | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, SID1 transmembrane family member 1, ZINC ION | | Authors: | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
7S0V
 
 | |
7RD0
 
 | |
7RCX
 
 | |
7RCW
 
 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCZ
 
 | | Crystal structure of C. difficile SpoVD in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCY
 
 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein, ZINC ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
3C5W
 
 | | Complex between PP2A-specific methylesterase PME-1 and PP2A core enzyme | | Descriptor: | PP2A A subunit, PP2A C subunit, PP2A-specific methylesterase PME-1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
3C5V
 
 | | PP2A-specific methylesterase apo form (PME) | | Descriptor: | Protein phosphatase methylesterase 1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
1T0B
 
 | | Structure of ThuA-like protein from Bacillus stearothermophilus | | Descriptor: | ThuA-like protein, ZINC ION | | Authors: | Borek, D, Chen, Y, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-08 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of chemical interactions in ThuA-like protein from Bacillus Stearothermophilus
To be Published
|
|
1T5Y
 
 | | Crystal Structure of Northeast Structural Genomics Consortium Target HR2118: A Human Homolog of Saccharomyces cerevisiae Nip7p | | Descriptor: | SACCHAROMYCES CEREVISIAE Nip7p HOMOLOG | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Acton, T.B, Shastry, R, Ma, L.-C, Cooper, B, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-05 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Northeast Structural Genomics Consortium Target HR2118:
A Human Homolog of Saccharomyces cerevisiae Nip7p
To be Published
|
|
4Q47
 
 | |
4ZOU
 
 | |
8KGR
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (32-MER), DNA (33-MER), DNA topoisomerase 2, ... | | Authors: | Cong, J, Xin, U, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of African swine fever virus topoisomerase II in complex with dsDNA
To Be Published
|
|
4Q48
 
 | |
1RYU
 
 | | Solution Structure of the SWI1 ARID | | Descriptor: | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1 | | Authors: | Kim, S, Zhang, Z, Upchurch, S, Isern, N, Chen, Y. | | Deposit date: | 2003-12-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition.
J.Biol.Chem., 279, 2004
|
|
5A9V
 
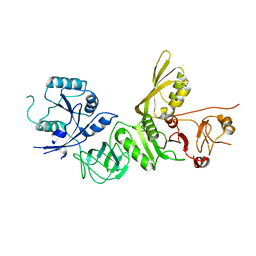 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9W
 
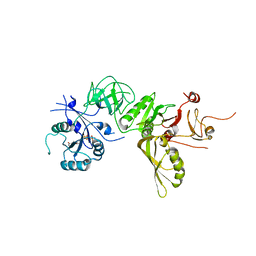 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1TZ9
 
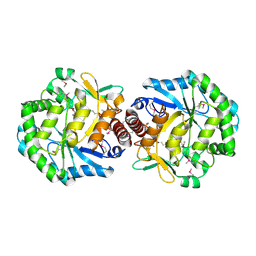 | | Crystal Structure of the Putative Mannonate Dehydratase from Enterococcus faecalis, Northeast Structural Genomics Target EfR41 | | Descriptor: | Mannonate dehydratase | | Authors: | Forouhar, F, Chen, Y, Xiao, R, Cooper, B, Shastry, R, Acton, T.A, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-07-09 | | Release date: | 2004-07-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Putative Mannonate Dehydratase from Enterococcus faecalis, Northeast Structural Genomics Target EfR41
To be Published
|
|
1U9C
 
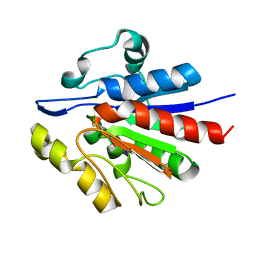 | | Crystallographic structure of APC35852 | | Descriptor: | APC35852 | | Authors: | Borek, D, Chen, Y, Shao, D, Collart, F, Joachimiak, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of DJI superfamily
To be Published
|
|
