6FM9
 
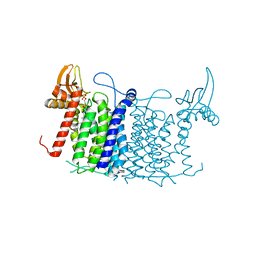 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6G46
 
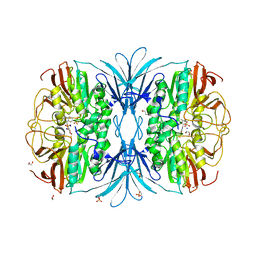 | |
6GGS
 
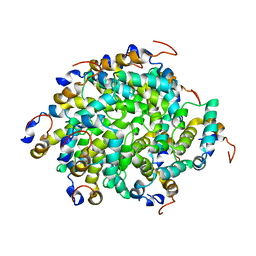 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6GFJ
 
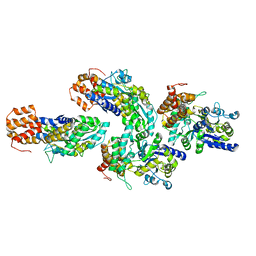 | | Structure of RIP2 CARD domain fused to crystallisable MBP tag | | Descriptor: | Sugar ABC transporter substrate-binding protein,Receptor-interacting serine/threonine-protein kinase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2018-04-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
8SSH
 
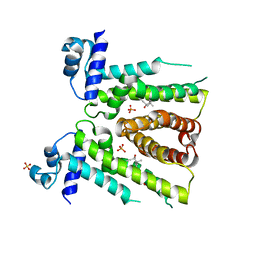 | | MtrR from Neisseria gonorrhoeae bound to Ethinyl Estradiol | | Descriptor: | Ethinyl estradiol, HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | Authors: | Hooks, G.M, Brennan, R.G. | | Deposit date: | 2023-05-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hormonal steroids induce multidrug resistance and stress response genes in Neisseria gonorrhoeae by binding to MtrR.
Nat Commun, 15, 2024
|
|
8SVI
 
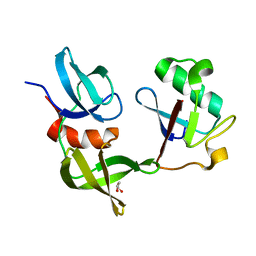 | | Ubiquitin variant i53:Mutant L67H with 53BP1 Tudor domain | | Descriptor: | GLYCEROL, Tumor protein p53 binding protein 1, Ubiquitin Variant i53: Mutant L67H | | Authors: | Partridge, J.R, Holden, J.K, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8SVJ
 
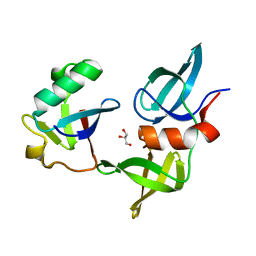 | | Ubiquitin variant i53: mutant VHH with 53BP1 Tudor domain | | Descriptor: | GLYCEROL, Tumor protein p53 binding protein 1, Ubiquitin varient i53 mutant VHH | | Authors: | Holden, J, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8SVG
 
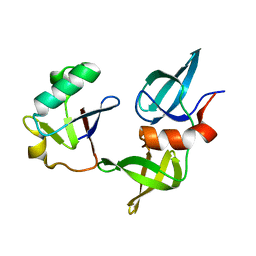 | | Ubiquitin variant i53 in complex with 53BP1 Tudor domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53 | | Authors: | Holden, J.K, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
8SVH
 
 | | Ubiquitin variant i53 mutant L67R bound to 53BP1 Tudor Domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53: mutant L67R | | Authors: | Holden, J.K, Partridge, J.R, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
7JUV
 
 | | Crystal Structure of KSR2:MEK1 in complex with AMP-PNP, and allosteric MEK inhibitor APS-9-95-1 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 2, MAGNESIUM ION, ... | | Authors: | Khan, Z.M, Dar, A.C, Scopton, A.P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural basis for the action of the drug trametinib at KSR-bound MEK.
Nature, 588, 2020
|
|
7JUT
 
 | |
6H9X
 
 | | Klebsiella pneumoniae Seryl-tRNA Synthetase in Complex with the Intermediate Analog 5'-O-(N-(L-Seryl)-Sulfamoyl)Adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CALCIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acylated sulfonamide adenosines as potent inhibitors of the adenylate-forming enzyme superfamily.
Eur.J.Med.Chem., 174, 2019
|
|
7JUQ
 
 | | Crystal Structure of KSR2:MEK1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 2, ... | | Authors: | Khan, Z.M, Dar, A.C. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the action of the drug trametinib at KSR-bound MEK.
Nature, 588, 2020
|
|
7JUR
 
 | |
7JUX
 
 | |
7JUU
 
 | |
7JUS
 
 | |
7JV0
 
 | |
7JUW
 
 | | Crystal Structure of KSR1:MEK1 in complex with AMP-PNP | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 1, MAGNESIUM ION, ... | | Authors: | Khan, Z.M, Dar, A.C. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis for the action of the drug trametinib at KSR-bound MEK.
Nature, 588, 2020
|
|
7JV1
 
 | | Crystal Structure of KSR1:MEK1 in complex with AMP-PNP, and allosteric MEK inhibitor APS-9-95-1 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 1, MAGNESIUM ION, ... | | Authors: | Khan, Z.M, Dar, A.C, Scopton, A.P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural basis for the action of the drug trametinib at KSR-bound MEK.
Nature, 588, 2020
|
|
7JUZ
 
 | |
7JUY
 
 | |
8T2D
 
 | | Ubiquitin variant i53:Mutant T12Y.T14E.L67R with 53BP1 Tudor domain | | Descriptor: | Tumor protein p53 binding protein 1, Ubiquitin variant i53 | | Authors: | Partridge, J.R, Holden, J.K, Wibowo, A.S, Mulichak, A. | | Deposit date: | 2023-06-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Functional screening in human HSPCs identifies optimized protein-based enhancers of Homology Directed Repair.
Nat Commun, 15, 2024
|
|
