1X65
 
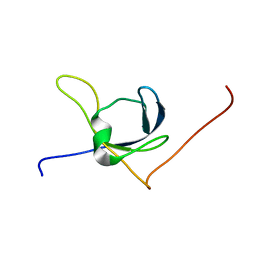 | | Solution structure of the third cold-shock domain of the human KIAA0885 protein (UNR PROTEIN) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
3G3E
 
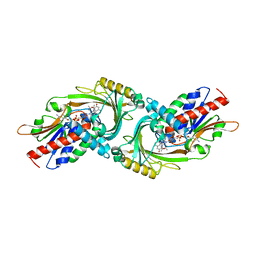 | |
7QWL
 
 | |
7QVC
 
 | |
7QVF
 
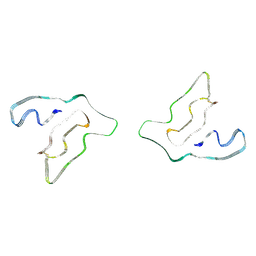 | |
7QWG
 
 | |
7QWM
 
 | |
7C3G
 
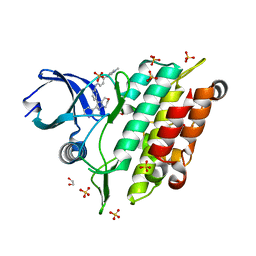 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with a bicyclic pyrazole inhibitor RK-73134 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-1, SULFATE ION, ... | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Novel bicyclic pyrazoles as potent ALK2 (R206H) inhibitors for the treatment of fibrodysplasia ossificans progressiva.
Bioorg.Med.Chem.Lett., 38, 2021
|
|
7FBR
 
 | |
6G52
 
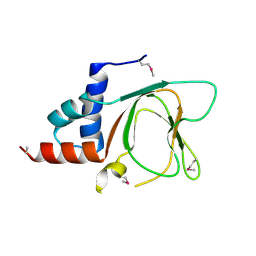 | | CRYSTAL STRUCTURE OF THE CNMP BINDING DOMAIN OF THE MAGNESIUM TRANSPORTER CNNM4 | | Descriptor: | Metal transporter CNNM4 | | Authors: | Gimenez, P, Oyenarte, I, Hardy, S, Zubillaga, M, Merino, N, Blanco, F.J, Siliqi, D, Tremblay, M, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2018-03-28 | | Release date: | 2019-04-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.691 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
1AHH
 
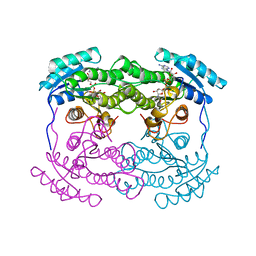 | | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEXED WITH NAD+ | | Descriptor: | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-08-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
6O3G
 
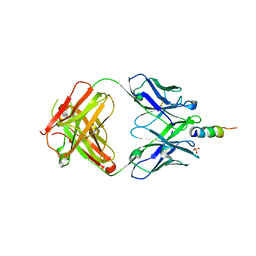 | |
6JUX
 
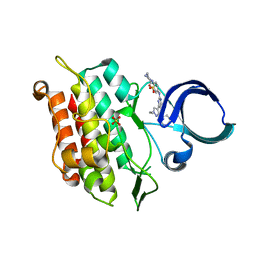 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with RK-71807 | | Descriptor: | 4-(1-ethyl-3-pyridin-3-yl-pyrazol-4-yl)-~{N}-(4-piperazin-1-ylphenyl)pyrimidin-2-amine, Activin receptor type-1, SULFATE ION | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Activin Receptor-Like Kinase 2 (R206H) Inhibition by Bis-heteroaryl Pyrazole-Based Inhibitors for the Treatment of Fibrodysplasia Ossificans Progressiva Identified by the Integration of Ligand-Based and Structure-Based Drug Design Approaches.
Acs Omega, 5, 2020
|
|
6O3K
 
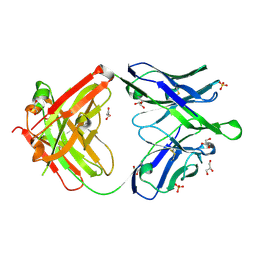 | |
6O3U
 
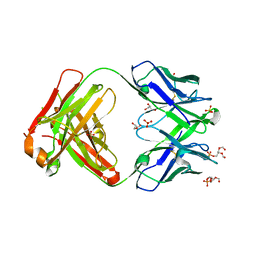 | |
6O3D
 
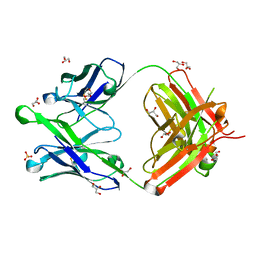 | |
6O3J
 
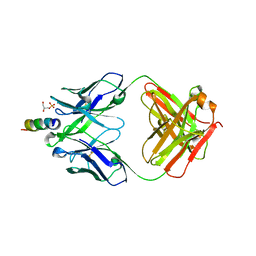 | | Crystal structure of the Fab fragment of the human HIV-1 neutralizing antibody PGZL1 in complex with its MPER peptide epitope (region 671-683 of HIV-1 gp41) and phosphatidic acid (06:0 PA) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, MPER peptide, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2019-02-26 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.416 Å) | | Cite: | An MPER antibody neutralizes HIV-1 using germline features shared among donors.
Nat Commun, 10, 2019
|
|
6O3L
 
 | |
6O41
 
 | |
1AHI
 
 | | 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE COMPLEXED WITH NADH AND 7-OXO GLYCOCHENODEOXYCHOLIC ACID | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 7 ALPHA-HYDROXYSTEROID DEHYDROGENASE, GLYCOCHENODEOXYCHOLIC ACID | | Authors: | Tanaka, N, Nonaka, T, Mitsui, Y. | | Deposit date: | 1995-08-25 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the binary and ternary complexes of 7 alpha-hydroxysteroid dehydrogenase from Escherichia coli.
Biochemistry, 35, 1996
|
|
6RS2
 
 | | Structure of the Bateman module of human CNNM4. | | Descriptor: | Metal transporter CNNM4 | | Authors: | Corral-Rodriguez, M.A, Stuiver, M, Gomez-Garcia, I, Oyenarte, I, Gimenez, P, Ereno-Orbea, J, Diercks, T, Muller, D, Martinez-Cruz, L.A. | | Deposit date: | 2019-05-21 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.694 Å) | | Cite: | Structural Insights into the Intracellular Region of the Human Magnesium Transport Mediator CNNM4.
Int J Mol Sci, 20, 2019
|
|
6O42
 
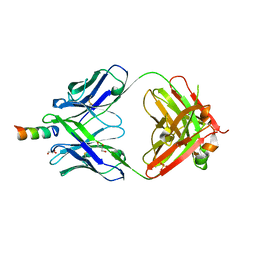 | |
3UMX
 
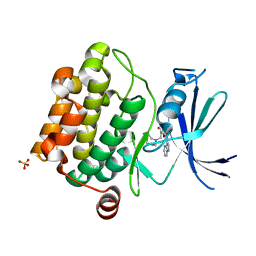 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indol-3-yl)methylene]-7-(azepan-1-ylmethyl)-6-hydroxybenzofuran-3(2H)-one | | Descriptor: | (2Z)-7-(azepan-1-ylmethyl)-6-hydroxy-2-(1H-indol-3-ylmethylidene)-1-benzofuran-3(2H)-one, Proto-oncogene serine/threonine-protein kinase pim-1, SULFATE ION | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7XJO
 
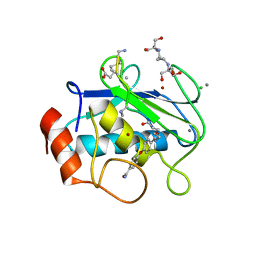 | | Crystal structure of human MMP-2 catalytic domain in complex with inhibitor | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kamitani, M, Mima, M, Takeuchi, T. | | Deposit date: | 2022-04-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aryloxyphenyl-Heptapeptide Hybrids as Potent and Selective Matrix Metalloproteinase-2 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis.
J.Med.Chem., 65, 2022
|
|
7XGJ
 
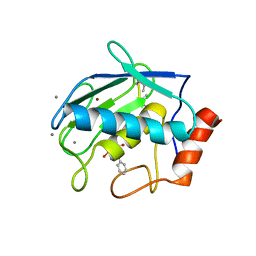 | | Crystal structure of human MMP-2 catalytic domain in complex with inhibitor | | Descriptor: | CALCIUM ION, GZS-ASN-ASP-ALA-LEU-IML-EOE-NH2, Matrix metalloproteinase-2, ... | | Authors: | Kamitani, M, Mima, M, Takeuchi, T. | | Deposit date: | 2022-04-05 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Aryloxyphenyl-Heptapeptide Hybrids as Potent and Selective Matrix Metalloproteinase-2 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis.
J.Med.Chem., 65, 2022
|
|
