5FO4
 
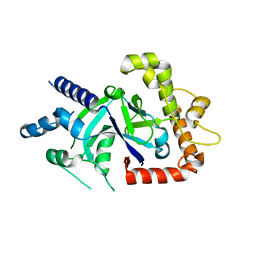 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P1) | | Descriptor: | LEUCYL TRNA SYNTHASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-18 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOC
 
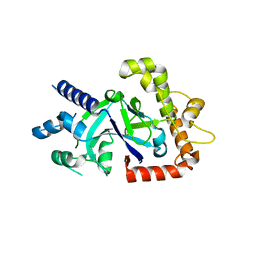 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P21) | | Descriptor: | LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOD
 
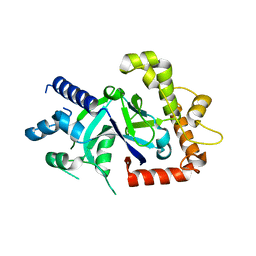 | | Crystal structure of the P.falciparum cytosolic leucyl-tRNA synthetase editing domain (space group P1) containing deletions of insertions 1 and 3 | | Descriptor: | 1,2-ETHANEDIOL, LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5FOF
 
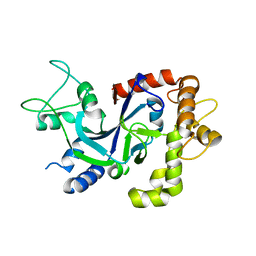 | | Crystal structure of the P.knowlesi cytosolic leucyl-tRNA synthetase editing domain | | Descriptor: | LEUCYL-TRNA SYNTHETASE | | Authors: | Palencia, A, Sonoiki, E, Guo, D, Ahyong, V, Dong, C, Li, X, Hernandez, V.S, Gut, J, Legac, J, Cooper, R, Alley, M.R.K, Freund, Y.R, DeRisi, J, Cusack, S, Rosenthal, P.J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anti-Malarial Benzoxaboroles Target P. Falciparum Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
2GO3
 
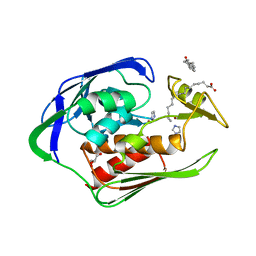 | | Crystal structure of Aquifex aeolicus LpxC complexed with imidazole. | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
3R27
 
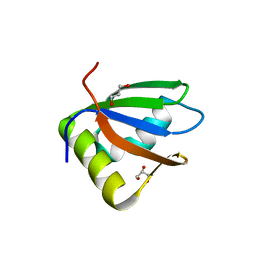 | | Crystal structure of the first RRM domain of heterogeneous nuclear ribonucleoprotein L (HnRNP L) | | Descriptor: | GLYCEROL, Heterogeneous nuclear ribonucleoprotein L | | Authors: | Zhang, W, Liu, Y, Zeng, F, Niu, L, Teng, M, Li, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the first RRM domain of heterogeneous nuclear ribonucleoprotein L (HnRNP L)
To be Published
|
|
2GVE
 
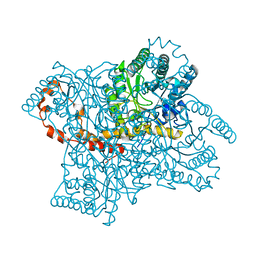 | | Time-of-Flight Neutron Diffraction Structure of D-Xylose Isomerase | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Katz, A.K, Li, X, Carrell, H.L, Hanson, B.L, Langan, P, Coates, L, Schoenborn, B.P, Glusker, J.P, Bunick, G.J. | | Deposit date: | 2006-05-02 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Locating active-site hydrogen atoms in D-xylose isomerase: Time-of-flight neutron diffraction.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GO4
 
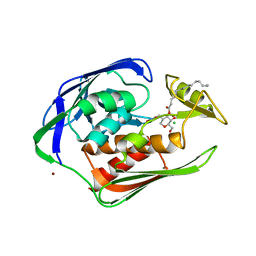 | | Crystal structure of Aquifex aeolicus LpxC complexed with TU-514 | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
7XY9
 
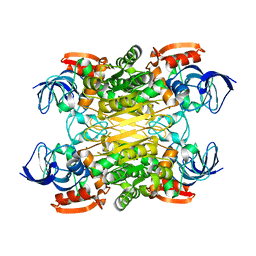 | | Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions | | Descriptor: | MAGNESIUM ION, NADP-dependent isopropanol dehydrogenase, ZINC ION | | Authors: | Chen, Q, Li, X, Yang, F, Qu, G, Sun, Z.T, Wang, Y.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions.
Nat Commun, 14, 2023
|
|
4EDL
 
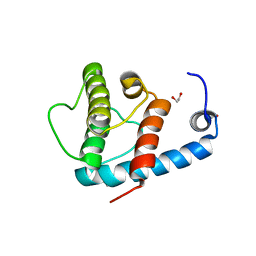 | | Crystal structure of beta-parvin CH2 domain | | Descriptor: | 1,2-ETHANEDIOL, Beta-parvin | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
4EDM
 
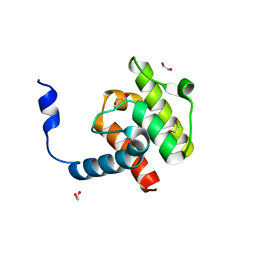 | | Crystal structure of beta-parvin CH2 domain | | Descriptor: | 1,2-ETHANEDIOL, Beta-parvin | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
8G05
 
 | | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | | Descriptor: | 6-(octylamino)pyrimidine-2,4(3H,5H)-dione, CHOLESTEROL, G-protein coupled receptor 84, ... | | Authors: | Zhang, X, Wang, Y.J, Li, X, Liu, G.B, Gong, W.M, Zhang, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-11-01 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Pro-phagocytic function and structural basis of GPR84 signaling.
Nat Commun, 14, 2023
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
7DCY
 
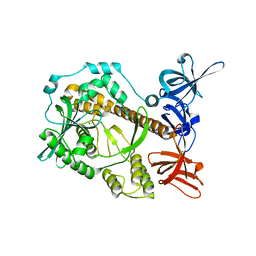 | | Apo form of Mycoplasma genitalium RNase R | | Descriptor: | MAGNESIUM ION, Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DIC
 
 | | Mycoplasma genitalium RNase R in complex with single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DOL
 
 | | Mycoplasma genitalium RNase R in complex with double-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
7DID
 
 | | Mycoplasma genitalium RNase R in complex with ribose methylated single-stranded RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*AP*AP*AP*A)-3'), Ribonuclease R | | Authors: | Abula, A, Quan, X, Li, X, Yang, T, Li, T, Chen, Q, Ji, X. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of RNase R substrate sensitivity for RNA ribose methylation.
Nucleic Acids Res., 49, 2021
|
|
4FQN
 
 | | Crystal structure of the CCM2 C-terminal Harmonin Homology Domain (HHD) | | Descriptor: | Malcavernin | | Authors: | Fisher, O.S, Zhang, R, Li, X, Murphy, J.W, Boggon, T.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of cerebral cavernous malformations 2 (CCM2) reveal a folded helical domain at its C-terminus.
Febs Lett., 587, 2013
|
|
4GFR
 
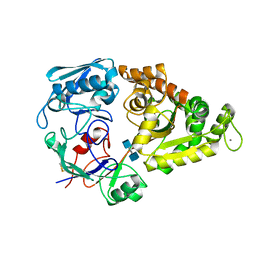 | | Crystal Structure of the liganded Chitin Oligasaccharide Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, Peptide ABC transporter, ... | | Authors: | Xu, S, Li, X, Gu, L, Roseman, R, Stock, A.M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chitin catabolic cascade in the marine bacterium Vibrio cholerae: properties, structure and functions of a periplasmic chitooligosaccharide binding protein (CBP)
To be Published
|
|
4EGL
 
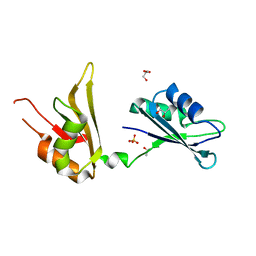 | | Crystal structure of two tandem RNA recognition motifs of Human antigen R | | Descriptor: | ELAV-like protein 1, GLYCEROL, SULFATE ION | | Authors: | Wang, H, Zeng, F, Liu, H, Teng, M, Li, X. | | Deposit date: | 2012-03-31 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of two tandem RNA recognition motifs of Human antigen R
To be Published
|
|
4FI1
 
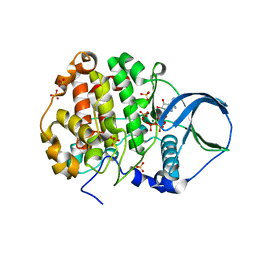 | | Crystal structure of scCK2 alpha in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Liu, H, Wang, H, Teng, M, Li, X. | | Deposit date: | 2012-06-07 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of scCK2 alpha in complex with ATP
To be Published
|
|
4DM4
 
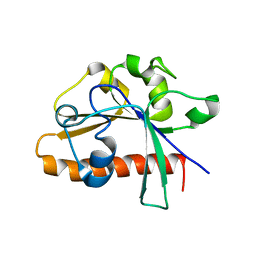 | | The conserved domain of yeast Cdc73 | | Descriptor: | Cell division control protein 73 | | Authors: | Chen, H, Shi, N, Gao, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4ED5
 
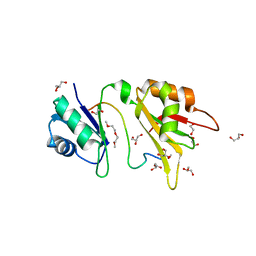 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | Authors: | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | Deposit date: | 2012-03-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4E7N
 
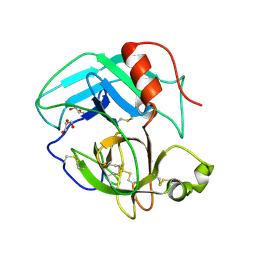 | | Crystal Structure of AhV_TL-I, a Glycosylated Snake-venom Thrombin-like Enzyme from Agkistrodon halys | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Snake-venom Thrombin-like Enzyme | | Authors: | Zeng, F, Li, X, Teng, M, Niu, L. | | Deposit date: | 2012-03-18 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of AhV_TL-I, a Glycosylated Snake-venom Thrombin-like Enzyme from Agkistrodon halys
to be published
|
|
6KNM
 
 | | Apelin receptor in complex with single domain antibody | | Descriptor: | Apelin receptor,Rubredoxin,Apelin receptor, Single domain antibody JN241, ZINC ION | | Authors: | Ma, Y.B, Ding, Y, Song, X, Ma, X, Li, X, Zhang, N, Song, Y, Sun, Y, Shen, Y, Zhong, W, Hu, L.A, Ma, Y.L, Zhang, M.Y. | | Deposit date: | 2019-08-06 | | Release date: | 2020-01-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-guided discovery of a single-domain antibody agonist against human apelin receptor.
Sci Adv, 6, 2020
|
|
