5WWW
 
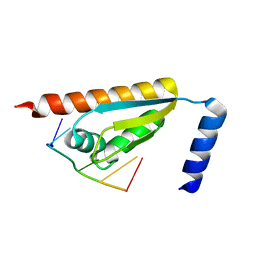 | | Crystal structure of the KH1 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*UP*UP*AP*G)-3'), RNA-binding E3 ubiquitin-protein ligase MEX3C | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
5WWZ
 
 | | Crystal structure of the KH2 domain of human RNA-binding E3 ubiquitin-protein ligase MEX-3C | | Descriptor: | RNA-binding E3 ubiquitin-protein ligase MEX3C, SULFATE ION | | Authors: | Yang, L, Wang, C, Li, F, Gong, Q. | | Deposit date: | 2017-01-05 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3-recognition element (MRE) motif with high affinity
J. Biol. Chem., 292, 2017
|
|
6ZZL
 
 | |
4C1W
 
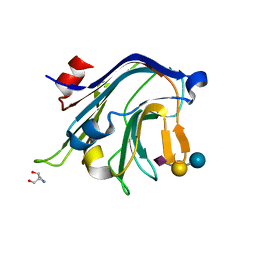 | | Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 3'-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, NEURAMINIDASE | | Authors: | Yang, L, Connaris, H, Potter, J.A, Taylor, G.L. | | Deposit date: | 2013-08-14 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Prevention of Influenza by Targeting Host Receptors Using Engineered Proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7EDQ
 
 | | MIF complex to compound7 | | Descriptor: | 6,7-bis(oxidanyl)chromen-2-one, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-03-16 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.266 Å) | | Cite: | Repurposing old drugs as novel inhibitors of human MIF from structural and functional analysis.
Bioorg.Med.Chem.Lett., 55, 2022
|
|
7EE8
 
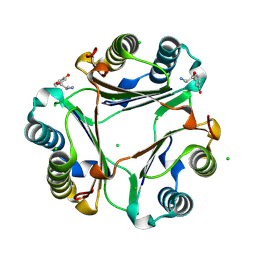 | | The crystal structure of MIF bound to compound D5 | | Descriptor: | 3-[(2R)-2-azanyl-1-oxidanyl-propyl]phenol, 3-[(2S)-2-azanyl-1-oxidanyl-propyl]phenol, CHLORIDE ION, ... | | Authors: | Fan, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-03-17 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Repurposing old drugs as novel inhibitors of human MIF from structural and functional analysis.
Bioorg.Med.Chem.Lett., 55, 2022
|
|
7E47
 
 | | Crystal structure of compound 6 bound to MIF | | Descriptor: | (2S,3R,4R,5R,6R)-2-[(2S,3R,4S,5R,6S)-6-(hydroxymethyl)-2-[[(2R,4R)-2-(4-hydroxyphenyl)-4,5-bis(oxidanyl)-3,4-dihydro-2H-chromen-7-yl]oxy]-4,5-bis(oxidanyl)oxan-3-yl]oxy-6-methyl-oxane-3,4,5-triol, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C.P, Yang, L, Guo, D.Y. | | Deposit date: | 2021-02-10 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
7E45
 
 | | Crystal structure of compound 7 bound to MIF | | Descriptor: | (2R)-7-[(2R,3R,4R,5S,6S)-6-(hydroxymethyl)-3-[(2R,3R,4R,5R,6S)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-(4-methoxy-3-oxidanyl-phenyl)-5-oxidanyl-2,3-dihydrochromen-4-one, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C.P, Yang, L. | | Deposit date: | 2021-02-10 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
7E4B
 
 | | Crystal structure of MIF bound to compound 5 | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
7E4C
 
 | | Crystal structure of MIF bound to compound11 | | Descriptor: | 4-[(3R)-8,8-dimethyl-3,4-dihydro-2H,8H-pyrano[2,3-f]chromen-3-yl]benzene-1,3-diol, CHLORIDE ION, Macrophage migration inhibitory factor, ... | | Authors: | Fan, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
7E4A
 
 | | Crystal structure of MIF bound to compound 13 | | Descriptor: | CHLORIDE ION, Flavoxate, Macrophage migration inhibitory factor, ... | | Authors: | Fa, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
7E49
 
 | | Crystal structure of MIF bound to compound10 | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Fan, C.P, Guo, D.Y, Yang, L. | | Deposit date: | 2021-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.574 Å) | | Cite: | Identification and Structure-Activity Relationships of Dietary Flavonoids as Human Macrophage Migration Inhibitory Factor (MIF) Inhibitors.
J.Agric.Food Chem., 69, 2021
|
|
9KTK
 
 | | Crystal structure of human SIRT3 with its activator SKLB-11A | | Descriptor: | MAGNESIUM ION, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Ouyang, L, Wu, C.Y. | | Deposit date: | 2024-12-02 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Unraveling Small Molecule-Mediated Sirtuin 3 Activation at a Distinct Binding Site for Cardioprotective Therapies.
Acs Cent.Sci., 11, 2025
|
|
6J92
 
 | |
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
5XEZ
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
5YQZ
 
 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4IAQ
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
6LN2
 
 | | Crystal structure of full length human GLP1 receptor in complex with Fab fragment (Fab7F38) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab7F38_heavy chain, Fab7F38_light chain, ... | | Authors: | Wu, F, Yang, L, Hang, K, Laursen, M, Wu, L, Han, G.W, Ren, Q, Roed, N.K, Lin, G, Hanson, M, Jiang, H, Wang, M, Reedtz-Runge, S, Song, G, Stevens, R.C. | | Deposit date: | 2019-12-28 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Full-length human GLP-1 receptor structure without orthosteric ligands.
Nat Commun, 11, 2020
|
|
4XYX
 
 | | NanB plus Optactamide | | Descriptor: | Optactamide, PHOSPHATE ION, Sialidase B | | Authors: | Rogers, G.W, Brear, P, Yang, L, Taylor, G.L, Westwood, N.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hunt for Serendipitous Allosteric Sites: Discovery of a novel allosteric inhibitor of the bacterial sialidase NanB
To Be Published
|
|
6NEF
 
 | | Outer Membrane Cytochrome S Filament from Geobacter Sulfurreducens | | Descriptor: | C-type cytochrome OmcS, HEME C, MAGNESIUM ION | | Authors: | Filman, D.J, Marino, S.F, Ward, J.E, Yang, L, Mester, Z, Bullitt, E, Lovley, D.R, Strauss, M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM reveals the structural basis of long-range electron transport in a cytochrome-based bacterial nanowire.
Commun Biol, 2, 2019
|
|
3P66
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | GOLD 3+ ION, Lysozyme C | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
