3SKR
 
 | | Crystal structure of the 2'- Deoxyguanosine riboswitch bound to 2'- Deoxyguanosine, cobalt Hexammine soak | | Descriptor: | 2'-DEOXY-GUANOSINE, COBALT HEXAMMINE(III), MAGNESIUM ION, ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
3SKZ
 
 | | Crystal structure of the 2'- deoxyguanosine riboswitch bound to guanosine | | Descriptor: | GUANOSINE, MAGNESIUM ION, RNA (68-MER), ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
2CW0
 
 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
2R19
 
 | |
4IB9
 
 | | Bovine beta-lactoglobulin (isoform B) in complex with dodecyltrimethylammonium (DTAC) | | Descriptor: | DODECANE-TRIMETHYLAMINE, GLYCEROL, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
4IB8
 
 | | Bovine beta-lactoglobulin (isoform A) in complex with dodecyl sulphate (SDS) | | Descriptor: | DODECYL SULFATE, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
4IB7
 
 | | Bovine beta-lactoglobulin (isoform A) in complex with dodecyltrimethylammonium (DTAC) | | Descriptor: | DODECANE-TRIMETHYLAMINE, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
4IBA
 
 | | Bovine beta-lactoglobulin (isoform B) in complex with dodecyl sulphate (SDS) | | Descriptor: | DODECYL SULFATE, GLYCEROL, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
5M3U
 
 | | The X-ray structure of human V216F phosphoglycerate kinase 1 mutant | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2016-10-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5MXM
 
 | | The X-ray structure of human M190I phosphoglycerate kinase 1 mutant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2017-01-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5M6Z
 
 | | The X-ray structure of human M189I PGK-1 mutant in partially closed conformation | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ilari, A, Fiorillo, A, Petrosino, M, Cipollone, A. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5M1R
 
 | | X-ray structure of human G166D PGK-1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoglycerate kinase 1 | | Authors: | Ilari, A, Cipollone, A, Fiorillo, A, Petrosino, M. | | Deposit date: | 2016-10-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5O7D
 
 | | The X-ray structure of human R38M phosphoglycerate kinase 1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoglycerate kinase 1 | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
2XHZ
 
 | | Probing the active site of the sugar isomerase domain from E. coli arabinose-5-phosphate isomerase via X-ray crystallography | | Descriptor: | ARABINOSE 5-PHOSPHATE ISOMERASE | | Authors: | Gourlay, L.J, Sommaruga, S, Nardini, M, Sperandeo, P, Deho, G, Polissi, A, Bolognesi, M. | | Deposit date: | 2010-06-24 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the Active Site of the Sugar Isomerase Domain from E. Coli Arabinose-5-Phosphate Isomerase Via X-Ray Crystallography.
Protein Sci., 19, 2010
|
|
1YKQ
 
 | | Crystal structure of Diels-Alder ribozyme | | Descriptor: | CADMIUM ION, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YKV
 
 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YLS
 
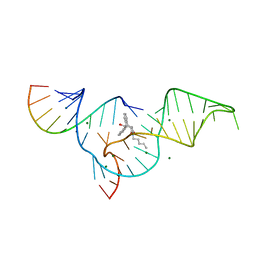 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
4IB6
 
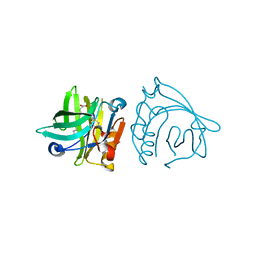 | | Bovine beta-lactoglobulin (isoform A) in complex with lauric acid (C12) | | Descriptor: | Beta-lactoglobulin, LAURIC ACID | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
6R7H
 
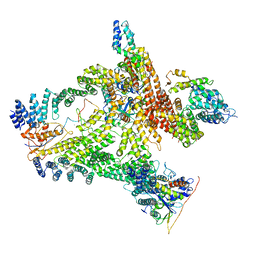 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7N
 
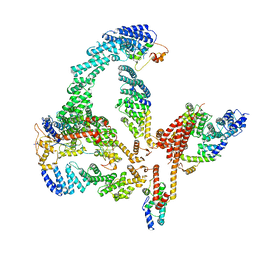 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7I
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
8VOX
 
 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgADP | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VP1
 
 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with ATP | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VOW
 
 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgATP and Vi | | Descriptor: | ABC-type bacteriocin transporter, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
8VOZ
 
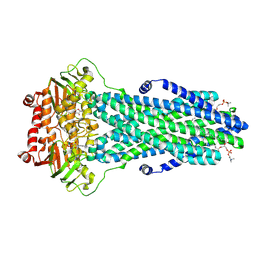 | | Cryo-EM structure of the ABC transporter PCAT1 cysteine-free core bound with MgADP and substrate | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxypropyl hexadecanoate, ABC-type bacteriocin transporter, ... | | Authors: | Zhang, R, Jagessar, K.L, Polasa, A, Brownd, M, Stein, R, Moradi, M, Karakas, E, Mchaourab, H.S. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Conformational cycle of a protease-containing ABC transporter in lipid nanodiscs reveals the mechanism of cargo-protein coupling.
Nat Commun, 15, 2024
|
|
