4UNC
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 8 DAYS INCUBATION IN 5MM MN (STATE 6) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNB
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 6 DAYS INCUBATION IN 5MM MN (STATE 5) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UN7
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA BEFORE INCUBATION IN 5MM MN (STATE 1) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, ZINC ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6QGS
 
 | | Crystal structure of APT1 bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, CHLORIDE ION, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
6QGQ
 
 | | Crystal structure of APT1 C2S mutant bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, GLYCEROL, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
6QGO
 
 | | Crystal structure of APT1 S119A mutant bound to palmitic acid. | | Descriptor: | Acyl-protein thioesterase 1, PALMITIC ACID | | Authors: | Audagnotto, M, Marcaida, M.J, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
2V07
 
 | | Structure of the Arabidopsis thaliana cytochrome c6A V52Q variant | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
2VMK
 
 | | Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain | | Descriptor: | RIBONUCLEASE E, SULFATE ION, ZINC ION | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
2V08
 
 | | Structure of wild-type Phormidium laminosum cytochrome c6 | | Descriptor: | CHLORIDE ION, CYTOCHROME C6, HEME C, ... | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
2VNF
 
 | | MOLECULAR BASIS OF HISTONE H3K4ME3 RECOGNITION BY ING4 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, HISTONE H3, ... | | Authors: | Palacios, A, Munoz, I.G, Pantoja-Uceda, D, Marcaida, M.J, Torres, D, Martin-Garcia, J.M, Luque, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular Basis of Histone H3K4Me3 Recognition by Ing4
J.Biol.Chem., 283, 2008
|
|
5AKF
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA NICKED IN THE CODING STRAND A AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*GP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5AKN
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA NICKED IN THE non-CODING STRAND B AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP *CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5AK9
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*GP)-3, ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
5AKM
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI G20S IN COMPLEX WITH ITS TARGET DNA IN THE PRESENCE OF 2MM MG | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
6HTF
 
 | |
7B1B
 
 | | Cryo-EM of Aedes Aegypti Toll5A dimer bound to Spz1C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAEL013433-PA, ... | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
7B1C
 
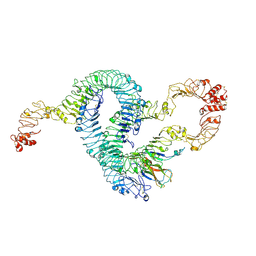 | | Cryo-EM of Aedes Aegypti Toll5A trimer bound to Spz1C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAEL013433-PA, ... | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
7B1D
 
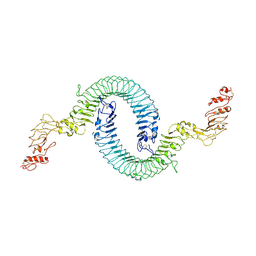 | | Cryo-EM of Aedes Aegypti Toll5A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
6RMH
 
 | | The Rigid-body refined model of the normal Huntingtin. | | Descriptor: | Huntingtin | | Authors: | Jung, T, Tamo, G, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2019-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
6YEJ
 
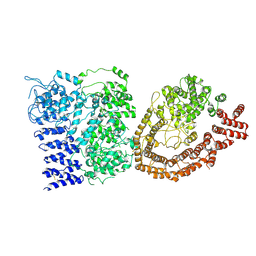 | | Cryo-EM structure of the Full-length disease type human Huntingtin | | Descriptor: | Huntingtin | | Authors: | Tame, G, Jung, T, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2020-03-24 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (18.200001 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
2KG4
 
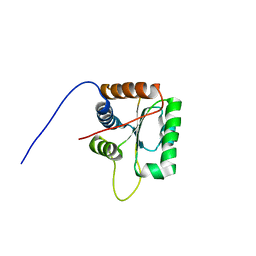 | | Three-dimensional structure of human Gadd45alpha in solution by NMR | | Descriptor: | Growth arrest and DNA-damage-inducible protein GADD45 alpha | | Authors: | Sanchez, R, Pantoja-Uceda, D, Prieto, J, Diercks, T, Campos-Olivas, R, Blanco, F.J. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase
J.Biol.Chem., 285, 2010
|
|
