5LBB
 
 | |
5L9R
 
 | |
5LBF
 
 | |
5L9B
 
 | |
5LAT
 
 | |
5LA9
 
 | |
5L9V
 
 | |
5LAS
 
 | |
4Y3B
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 201D with 14-3-3sigma | | Descriptor: | (2S)-2-(2-methoxyethyl)pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-THR-[H][C@@]1(C(C2=CC=CC=C2)C3=CC=CC=C3)CCCN1C | | Authors: | Bartel, M, Milroy, L.G, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-10 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4Y32
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 109B with 14-3-3sigma | | Descriptor: | (2S)-2-(2-methoxyethyl)pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-CNC(C(C)O)C(=O)N1CCCC1CCOC | | Authors: | Bartel, M, Milroy, L, Bier, D, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-10 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4Y5I
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 126B with 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Microtubule-associated protein tau | | Authors: | Leysen, S, Bartel, M, Milroy, L, Brunsveld, L, Ottmann, C. | | Deposit date: | 2015-02-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6FAU
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2e-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-(2-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FI4
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 3.2e with 14-3-3sigma | | Descriptor: | (2~{S})-2-(diphenylmethyl)pyrrolidine, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FAV
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FBY
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2b with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{S})-(3-methylphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-16 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FI5
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 3.2d with 14-3-3sigma | | Descriptor: | (2~{S})-2-(diphenylmethyl)pyrrolidine, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FAW
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2c-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-[2-(2-methoxyethoxy)phenyl]-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FBW
 
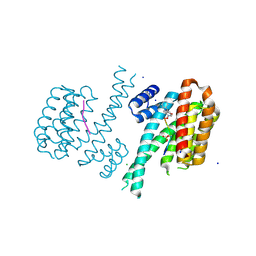 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-II with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{S})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6YW2
 
 | |
6YW3
 
 | |
6YW4
 
 | |
6YW1
 
 | |
6RK8
 
 | | Fragment AZ-014 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-(3-azanyl-4-methyl-pyrazol-1-yl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RL3
 
 | | Fragment AZ-003 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-azanyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RL6
 
 | | Fragment AZ-024 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
