6KW5
 
 | | The ClassC RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome
To Be Published
|
|
6MIZ
 
 | |
6N1H
 
 | | Cryo-EM structure of ASC-CARD filament | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MB2
 
 | | Cryo-EM structure of the PYD filament of AIM2 | | Descriptor: | Green fluorescent protein, Interferon-inducible protein AIM2 | | Authors: | Lu, A, Li, Y, Wu, H. | | Deposit date: | 2018-08-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2.
Cell Discov, 1, 2015
|
|
6MIX
 
 | | Human TRPM2 ion channel in apo state | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
6N1I
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MK7
 
 | | Solution structure of the large extracellular loop of FtsX in Streptococcus pneumoniae | | Descriptor: | Cell division protein FtsX | | Authors: | Edmonds, K.A, Fu, Y, Wu, H, Rued, B.E, Bruce, K.E, Winkler, M.E, Giedroc, D.P. | | Deposit date: | 2018-09-25 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
MBio, 10, 2019
|
|
6MJ2
 
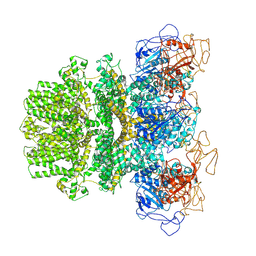 | | Human TRPM2 ion channel in a calcium- and ADPR-bound state | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.36 Å) | | Cite: | Structures and gating mechanism of human TRPM2.
Science, 362, 2018
|
|
6NCV
 
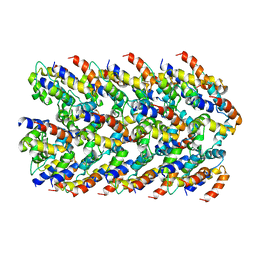 | | Cryo-EM structure of NLRP6 PYD filament | | Descriptor: | NACHT, LRR and PYD domains-containing protein 6 | | Authors: | Shen, C, Fu, T.M, Wu, H. | | Deposit date: | 2018-12-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism for NLRP6 inflammasome assembly and activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7DWO
 
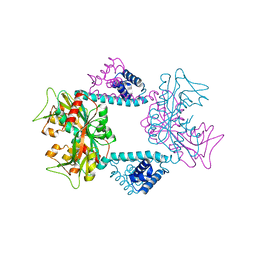 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | Authors: | Wang, W.W, Wu, H, He, J.H, Yu, F. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
6NDJ
 
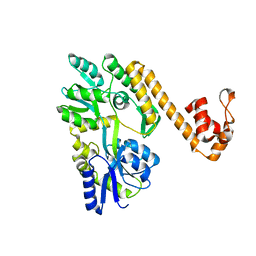 | | Crystal structure of human NLRP6 PYD domain with MBP fusion | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, NACHT, LRR and PYD domains-containing protein 6 chimera | | Authors: | Shen, C, Fu, T.M, Wu, H. | | Deposit date: | 2018-12-13 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular mechanism for NLRP6 inflammasome assembly and activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8P1S
 
 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) in complex with fucose. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819), ... | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
5XF8
 
 | | Cryo-EM structure of the Cdt1-MCM2-7 complex in AMPPNP state | | Descriptor: | Cell division cycle protein CDT1, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Zhai, Y, Cheng, E, Wu, H, Li, N, Yung, P.Y, Gao, N, Tye, B.K. | | Deposit date: | 2017-04-09 | | Release date: | 2017-05-03 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W0R
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cacodylic acid | | Descriptor: | CACODYLATE ION, CALCIUM ION, MBP fused activation-induced cytidine deaminase, ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W1C
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W0U
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W0Z
 
 | |
1DS9
 
 | | SOLUTION STRUCTURE OF CHLAMYDOMONAS OUTER ARM DYNEIN LIGHT CHAIN 1 | | Descriptor: | OUTER ARM DYNEIN | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2000-01-07 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a dynein motor domain associated light chain.
Nat.Struct.Biol., 7, 2000
|
|
4WD5
 
 | | Crystal structure of EGFR 696-1022 T790M in complex with QL-X138 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-{2-methyl-5-[2-oxo-9-(1H-pyrazol-4-yl)benzo[h][1,6]naphthyridin-1(2H)-yl]phenyl}propanamide | | Authors: | Yun, C.H, Eck, M.J. | | Deposit date: | 2014-09-07 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of EGFR 696-1022 T790M in complex with QL-X138
To Be Published
|
|
1M9L
 
 | | Relaxation-based Refined Structure Of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Outer Arm Dynein Light Chain 1 | | Authors: | Wu, H.W, Maciejewski, M.W, Marintchev, A, Benashski, S.E, Mullen, G.P, King, S.M. | | Deposit date: | 2002-07-29 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Relaxation-based structure refinement and backbone molecular dynamics of the
Dynein motor domain-associated light chain
Biochemistry, 42, 2003
|
|
4ZSE
 
 | |
2H11
 
 | | Amino-terminal Truncated Thiopurine S-Methyltransferase Complexed with S-Adenosyl-L-Homocysteine | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOCYANATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2006-05-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis of allele variation of human thiopurine-S-methyltransferase.
Proteins, 67, 2007
|
|
4C1Q
 
 | | Crystal structure of the PRDM9 SET domain in complex with H3K4me2 and AdoHcy. | | Descriptor: | GLYCEROL, HISTONE H3.1, HISTONE-LYSINE N-METHYLTRANSFERASE PRDM9, ... | | Authors: | Mathioudakis, N, Cusack, S, Kadlec, J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Regulation of the H3K4 Methyltransferase Activity of Prdm9.
Cell Rep., 5, 2013
|
|
2JOL
 
 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | Descriptor: | Putative G-nucleotide exchange factor | | Authors: | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | Deposit date: | 2007-03-14 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
