5FFR
 
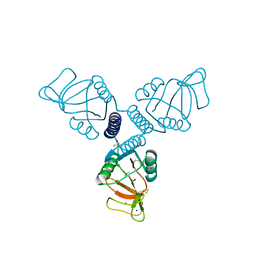 | | Crystal Structure of Surfactant Protein-A complexed with phosphocholine | | Descriptor: | CALCIUM ION, PHOSPHOCHOLINE, Pulmonary surfactant-associated protein A, ... | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
5H8O
 
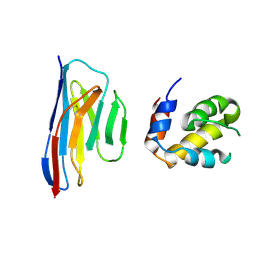 | | Crystal structure of an ASC-binding nanobody in complex with the CARD domain of ASC | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD, VHH nanobody | | Authors: | Lu, A, Schmidt, F.I, Ruan, J, Tang, C, Wu, H, Ploegh, H.L. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-06 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (4.206 Å) | | Cite: | A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly.
J.Exp.Med., 213, 2016
|
|
2PON
 
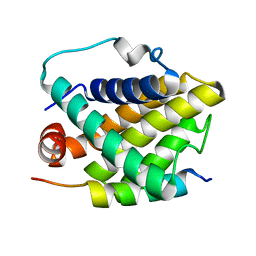 | | Solution structure of the Bcl-xL/Beclin-1 complex | | Descriptor: | Apoptosis regulator Bcl-X, Beclin-1 | | Authors: | Feng, W, Huang, S, Wu, H, Zhang, M. | | Deposit date: | 2007-04-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Bcl-xL's Target Recognition Versatility Revealed by the Structure of Bcl-xL in Complex with the BH3 Domain of Beclin-1.
J.Mol.Biol., 372, 2007
|
|
2PSW
 
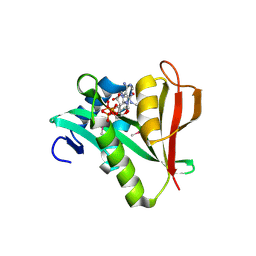 | | Human MAK3 homolog in complex with CoA | | Descriptor: | COENZYME A, N-acetyltransferase 13 | | Authors: | Walker, J.R, Schuetz, A, Antoshenko, T, Wu, H, Bernstein, G, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Human MAK3 homolog.
To be Published
|
|
2NS5
 
 | |
4QIM
 
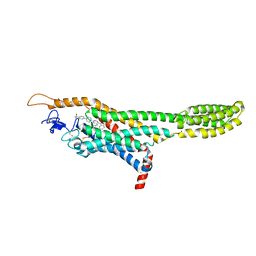 | | Structure of the human smoothened receptor in complex with ANTA XV | | Descriptor: | 2-{6-[4-(4-benzylphthalazin-1-yl)piperazin-1-yl]pyridin-3-yl}propan-2-ol, Smoothened homolog/Soluble cytochrome b562 chimeric protein, ZINC ION | | Authors: | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
2OB0
 
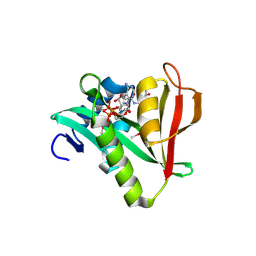 | | Human MAK3 homolog in complex with Acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Human MAK3 homolog | | Authors: | Walker, J.R, Schuetz, S, Antoshenko, T, Wu, H, Bernstein, G, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-18 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human MAK3 homolog
To be Published
|
|
4QIN
 
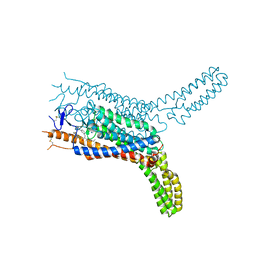 | | Structure of the human smoothened receptor in complex with SAG1.5 | | Descriptor: | 3-chloro-4,7-difluoro-N-[trans-4-(methylamino)cyclohexyl]-N-[3-(pyridin-4-yl)benzyl]-1-benzothiophene-2-carboxamide, Smoothened homolog/Soluble cytochrome b562 chimeric protein | | Authors: | Wang, C, Wu, H, Evron, T, Vardy, E, Han, G.W, Huang, X.-P, Hufeisen, S.J, Mangano, T.J, Urban, D.J, Katritch, V, Cherezov, V, Caron, M.G, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-05-31 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
4QL1
 
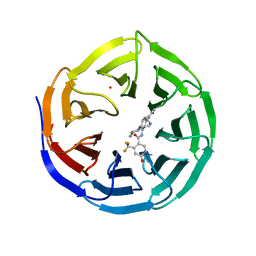 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
6K15
 
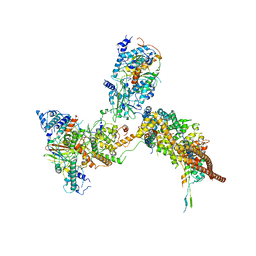 | | RSC substrate-recruitment module | | Descriptor: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
5H8D
 
 | | Crystal structure of an ASC binding nanobody | | Descriptor: | VHH nanobody | | Authors: | Lu, A, Schmidt, F.I, Ruan, J, Tang, C, Wu, H, Ploegh, H.L. | | Deposit date: | 2015-12-23 | | Release date: | 2016-04-06 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A single domain antibody fragment that recognizes the adaptor ASC defines the role of ASC domains in inflammasome assembly.
J.Exp.Med., 213, 2016
|
|
2QL1
 
 | | Structural Characterization of a Mutated, ADCC-Enhanced Human Fc Fragment | | Descriptor: | IGHM protein, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.534 Å) | | Cite: | Structural characterization of a mutated, ADCC-enhanced human Fc fragment
Mol.Immunol., 45, 2008
|
|
4RWS
 
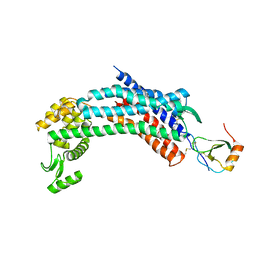 | | Crystal structure of CXCR4 and viral chemokine antagonist vMIP-II complex (PSI Community Target) | | Descriptor: | C-X-C chemokine receptor type 4/Endolysin chimeric protein, Viral macrophage inflammatory protein 2 | | Authors: | Qin, L, Kufareva, I, Holden, L, Wang, C, Zheng, Y, Wu, H, Fenalti, G, Han, G.W, Cherezov, V, Abagyan, R, Stevens, R.C, Handel, T.M, GPCR Network (GPCR) | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine.
Science, 347, 2015
|
|
3QAK
 
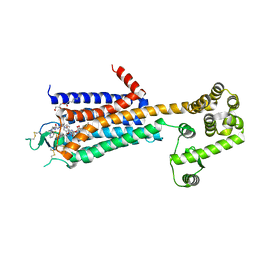 | | Agonist bound structure of the human adenosine A2a receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Adenosine receptor A2a,lysozyme chimera | | Authors: | Xu, F, Wu, H, Katritch, V, Han, G.W, Cherezov, V, Stevens, R, GPCR Network (GPCR) | | Deposit date: | 2011-01-11 | | Release date: | 2011-03-09 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an agonist-bound human A2A adenosine receptor.
Science, 332, 2011
|
|
4N0F
 
 | |
4L5R
 
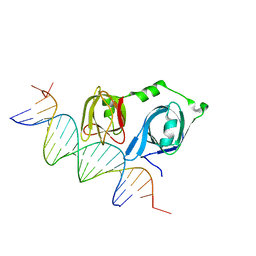 | |
6BFN
 
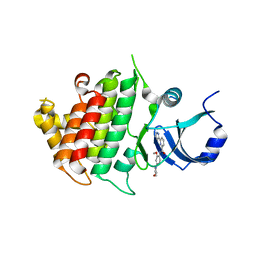 | | Crystal structure of human IRAK1 | | Descriptor: | Interleukin-1 receptor-associated kinase 1, N-[2-methoxy-4-(morpholin-4-yl)phenyl]-6-(1H-pyrazol-5-yl)pyridine-2-carboxamide | | Authors: | Wang, L, Qiao, Q, Wu, H. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human IRAK1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4L5Q
 
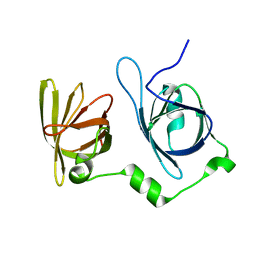 | | Crystal structure of p202 HIN1 | | Descriptor: | Interferon-activable protein 202 | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
4LCT
 
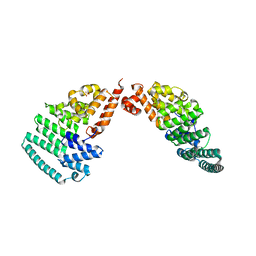 | | Crystal Structure and Versatile Functional Roles of the COP9 Signalosome Subunit 1 | | Descriptor: | COP9 signalosome complex subunit 1, SULFATE ION | | Authors: | Lee, J.-H, Yi, L, Li, J, Schweitzer, K, Borgmann, M, Naumann, M, Wu, H. | | Deposit date: | 2013-06-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and versatile functional roles of the COP9 signalosome subunit 1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4N0U
 
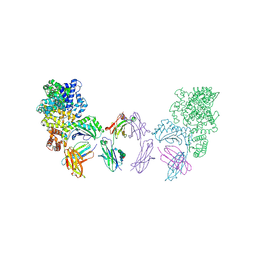 | | Ternary complex between Neonatal Fc receptor, serum albumin and Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Ig gamma-1 chain C region, ... | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2013-10-02 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Insights into Neonatal Fc Receptor-based Recycling Mechanisms.
J.Biol.Chem., 289, 2014
|
|
4LD6
 
 | | PWWP domain of human PWWP Domain-Containing Protein 2B | | Descriptor: | GLYCEROL, PWWP domain-containing protein 2B, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dombrovski, L, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PWWP domain of human PWWP Domain-Containing Protein 2B
To be Published
|
|
6BZE
 
 | | Cryo-EM structure of BCL10 CARD filament | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | David, L, Li, Y, Ma, J, Garner, E, Zhang, X, Wu, H. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Assembly mechanism of the CARMA1-BCL10-MALT1-TRAF6 signalosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4L5S
 
 | | p202 HIN1 in complex with 12-mer dsDNA | | Descriptor: | 12-mer DNA, Interferon-activable protein 202, SULFATE ION | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
4L5T
 
 | | Crystal structure of the tetrameric p202 HIN2 | | Descriptor: | Interferon-activable protein 202 | | Authors: | Yin, Q, Tian, Y, Wu, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Molecular Mechanism for p202-Mediated Specific Inhibition of AIM2 Inflammasome Activation.
Cell Rep, 4, 2013
|
|
4LWD
 
 | | Human CARMA1 CARD domain | | Descriptor: | Caspase recruitment domain-containing protein 11, MAGNESIUM ION, SULFATE ION | | Authors: | Zheng, C, Wu, H. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly.
Mol.Cell, 51, 2013
|
|
