5EDM
 
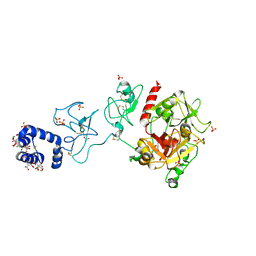 | | Crystal structure of prothrombin deletion mutant residues 154-167 ( Form I ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
5EE3
 
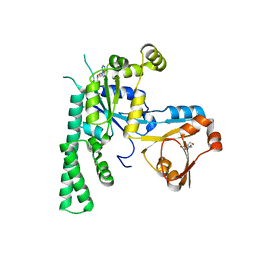 | | COMPLEX STRUCTURE OF OSYCHF1 WITH AMP-PNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EE0
 
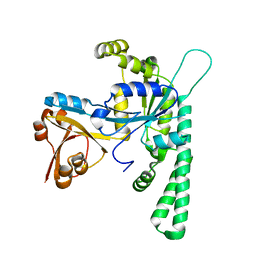 | | Crystal structure of OsYchF1 at pH 6.5 | | Descriptor: | Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8XQ3
 
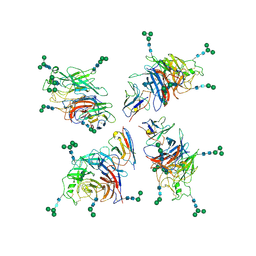 | | Structure of Nipah virus Bangladesh string G protein ectodomain tetramer bound to single-domain antibody n425 at 5.87 Angstroms overall resolution | | Descriptor: | Nipah virus Bangladesh string G protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, L, Chen, Z, Sun, Y, Mao, Q. | | Deposit date: | 2024-01-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.87 Å) | | Cite: | Fully human single-domain antibody targeting a highly conserved cryptic epitope on the Nipah virus G protein.
Nat Commun, 15, 2024
|
|
8XPY
 
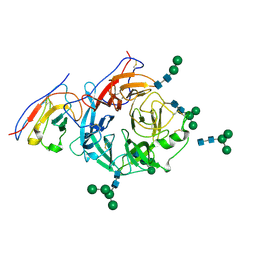 | | Structure of Nipah virus Malaysia string G protein ectodomain monomer bound to single-domain antibody n425 at 3.63 Angstroms overall resolution | | Descriptor: | Glycoprotein G, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, L, Chen, Z, Sun, Y, Mao, Q. | | Deposit date: | 2024-01-04 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Fully human single-domain antibody targeting a highly conserved cryptic epitope on the Nipah virus G protein.
Nat Commun, 15, 2024
|
|
8XPS
 
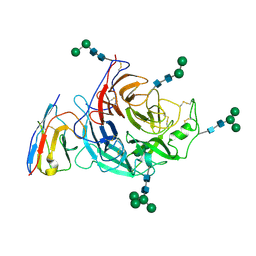 | | Structure of Nipah virus Bangladesh string G protein ectodomain monomer bound to single-domain antibody n425 at 3.22 Angstroms overall resolution | | Descriptor: | Nipah virus Bangladesh string G protein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, L, Chen, Z, Sun, Y, Mao, Q. | | Deposit date: | 2024-01-04 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Fully human single-domain antibody targeting a highly conserved cryptic epitope on the Nipah virus G protein.
Nat Commun, 15, 2024
|
|
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
5YUF
 
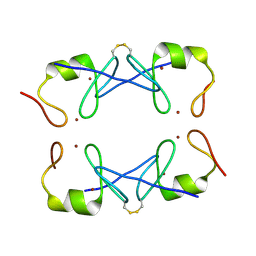 | | Crystal Structure of PML RING tetramer | | Descriptor: | Protein PML, ZINC ION | | Authors: | Wang, P, Benhend, S, Wu, H, Breitenbach, V, Zhen, T, Jollivet, F, Peres, L, Li, Y, Chen, S, Chen, Z, de THE, H, Meng, G. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RING tetramerization is required for nuclear body biogenesis and PML sumoylation.
Nat Commun, 9, 2018
|
|
5Z2T
 
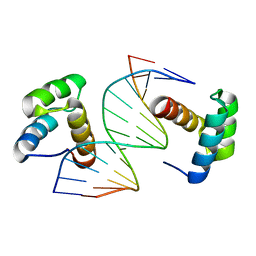 | | Crystal structure of DNA-bound DUX4-HD2 | | Descriptor: | 5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*TP*T)-3', 5'-D(P*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3', Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.623 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
4GEE
 
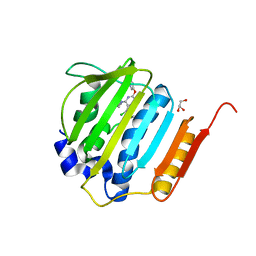 | | Pyrrolopyrimidine inhibitors of DNA gyrase B and topoisomerase IV, part I: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-yloxy)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Chen, Z, Tari, L.W. | | Deposit date: | 2012-08-01 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4FTS
 
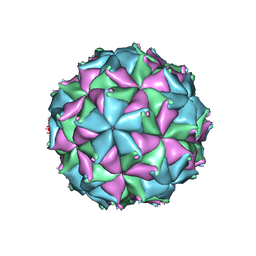 | | Crystal structure of the N363T mutant of the Flock House virus capsid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
8HB2
 
 | |
8HBB
 
 | |
8HAZ
 
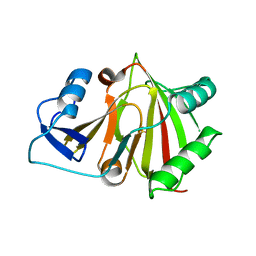 | |
4FSJ
 
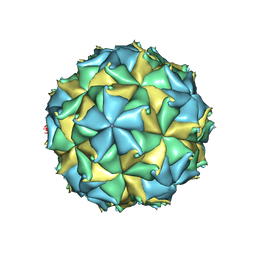 | | Crystal structure of the virus like particle of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4FTE
 
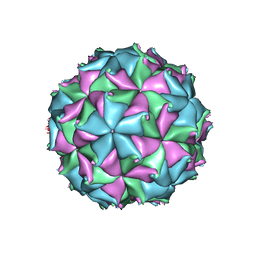 | | Crystal structure of the D75N mutant capsid of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4FTB
 
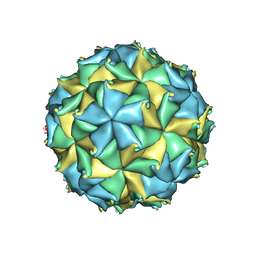 | | Crystal structure of the authentic Flock House virus particle | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
4HFP
 
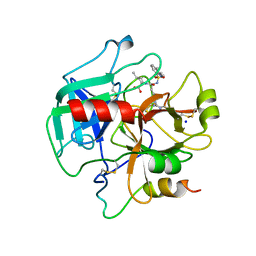 | | Structure of thrombin mutant S195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Prothrombin, SODIUM ION | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Lin, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-10-05 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
7VJV
 
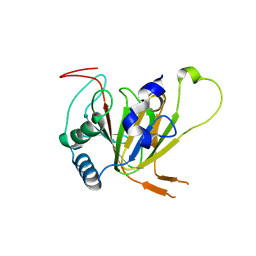 | | Human AlkB homolog ALKBH6 in complex with alpha-katoglutarate and Mn | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, MANGANESE (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VJS
 
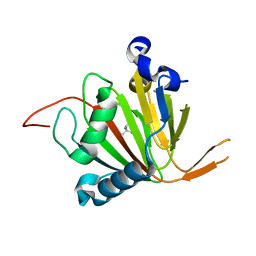 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7YL2
 
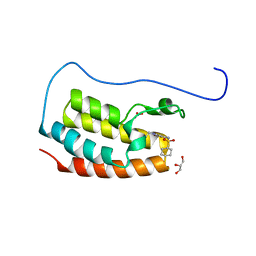 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | Authors: | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-07-25 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
3R3G
 
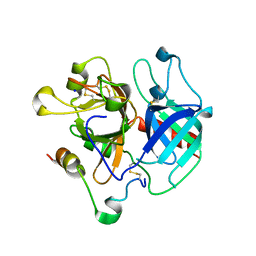 | | Structure of human thrombin with residues 145-150 of murine thrombin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin Heavy Chain, ... | | Authors: | Pozzi, N, Chen, R, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rigidification of the autolysis loop enhances Na(+) binding to thrombin.
Biophys.Chem., 159, 2011
|
|
7VVZ
 
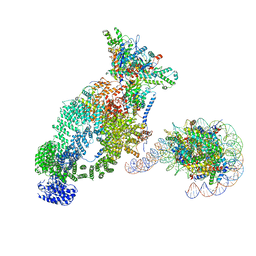 | | NuA4 bound to the nucleosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Qu, K, Chen, Z. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
8GQ6
 
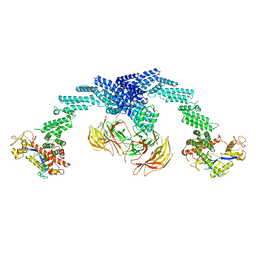 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
