6VG3
 
 | |
6SBA
 
 | | Crystal Structure of mTEAD with a VGL4 Tertiary Structure Mimetic | | Descriptor: | ACETYL GROUP, GLYCEROL, Transcriptional enhancer factor TEF-3, ... | | Authors: | Adihou, H, Grossmann, T.N, Waldmann, H, Gasper, R. | | Deposit date: | 2019-07-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A protein tertiary structure mimetic modulator of the Hippo signalling pathway.
Nat Commun, 11, 2020
|
|
7SVN
 
 | | DPP9 IN COMPLEX WITH LIGAND ICeD-1 | | Descriptor: | (2S,4S)-1-[(2S)-2-amino-2-cyclohexylacetyl]-4-fluoropyrrolidine-2-carbonitrile, Dipeptidyl peptidase 9 | | Authors: | Lammens, A, Hollenstein, K, Klein, D.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A Phenotypic Screen Identifies Potent DPP9 Inhibitors Capable of Killing HIV-1 Infected Cells.
Acs Chem.Biol., 17, 2022
|
|
6X3P
 
 | | Co-structure of BTK kinase domain with L-005298385 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{8-amino-3-[(6R,8aS)-3-oxooctahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-(cyclopropyloxy)-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6TU9
 
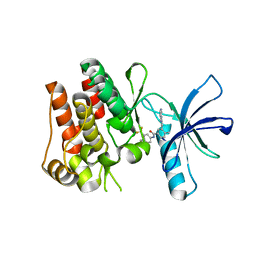 | | The ROR1 Pseudokinase Domain Bound To Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Niininen, W, Ungureanu, D, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
6TUA
 
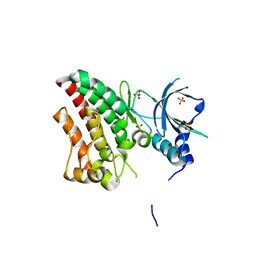 | | The RYK Pseudokinase Domain | | Descriptor: | SULFATE ION, Tyrosine-protein kinase RYK | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
5O8M
 
 | |
4R6R
 
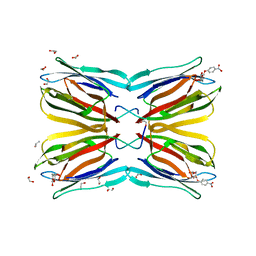 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6Q
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5HD7
 
 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Mutant SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5HD4
 
 | | Dissecting Therapeutic Resistance to ERK Inhibition Rat Wild Type SCH772984 in complex with (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Jha, S, Morris, E.J, Hruza, A, Mansueto, M.S, Schroeder, G, Arbanas, J, McMasters, D, Restaino, C.R, Dayananth, R, Black, S, Elsen, N.L, Mannarino, A, Cooper, A, Fawell, S, Zawel, L, Jayaraman, L, Samatar, A.A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dissecting Therapeutic Resistance to ERK Inhibition.
Mol.Cancer Ther., 15, 2016
|
|
5ZFO
 
 | | NMR structure of IRD12 from Capsicum annum. | | Descriptor: | Pin-II type proteinase inhibitor 38 | | Authors: | Gartia, J, Barnwal, R.P, Chary, K.V.R. | | Deposit date: | 2018-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of inhibitory repeat domain variant 12, a plant protease inhibitor from Capsicum annuum, and its structural relationship to other plant protease inhibitors.
J.Biomol.Struct.Dyn., 2019
|
|
4PXI
 
 | | Elucidation of the Structural and Functional Mechanism of Action of the TetR Family Protein, CprB from S. coelicolor A3(2) | | Descriptor: | CprB, DNA (5'-D(*AP*CP*AP*TP*AP*CP*GP*GP*GP*AP*CP*GP*CP*CP*CP*CP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*CP*GP*GP*GP*GP*CP*GP*TP*CP*CP*CP*GP*TP*AP*TP*GP*T)-3') | | Authors: | Hussain, B, Ruchika, B, Aruna, B, Ruchi, A. | | Deposit date: | 2014-03-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2)
Nucleic Acids Res., 42, 2014
|
|
6PCB
 
 | |
4OS5
 
 | |
6XVU
 
 | |
4OS1
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK601 (bicyclic 1) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS6
 
 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK604 (bicyclic 2) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
6PCC
 
 | |
7CFD
 
 | | Drosophila melanogaster Krimper eTud2-AubR15me2 complex | | Descriptor: | FI20010p1, Protein aubergine | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFB
 
 | | Drosophila melanogaster Krimper eTud1 apo structure | | Descriptor: | FI20010p1, SULFATE ION | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
7CFC
 
 | | Drosophila melanogaster Krimper eTud1-Ago3 complex | | Descriptor: | FI20010p1, Protein argonaute-3 | | Authors: | Hu, H, Li, S. | | Deposit date: | 2020-06-25 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex.
Nat Commun, 12, 2021
|
|
