6MVO
 
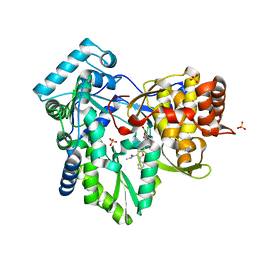 | | HCV NS5B 1A Y316 bound to Compound 49 | | Descriptor: | 6-[(7-chloro-1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)(methylsulfonyl)amino]-5-cyclopropyl-2-(4-fluorophenyl)-N-methyl-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
3DQR
 
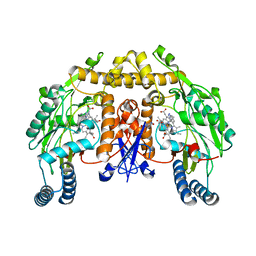 | | Structure of neuronal NOS D597N/M336V mutant heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
3B3P
 
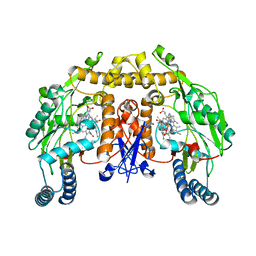 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-(3-chlorobenzyl)ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
7UOV
 
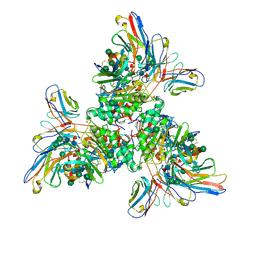 | |
7UOT
 
 | | Native Lassa glycoprotein in complex with neutralizing antibodies 8.9F and 37.2D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 37.2D heavy chain (variable domain), ... | | Authors: | Li, H, Saphire, E.O. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | A cocktail of protective antibodies subverts the dense glycan shield of Lassa virus.
Sci Transl Med, 14, 2022
|
|
3QSD
 
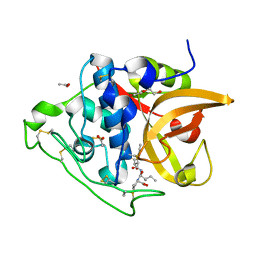 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with CA074 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-02-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
7JLT
 
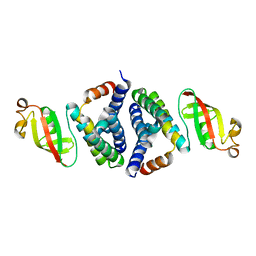 | | Crystal Structure of SARS-CoV-2 NSP7-NSP8 complex. | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Biswal, M, Hai, R, Song, J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP.
Nucleic Acids Res., 49, 2021
|
|
3S3Q
 
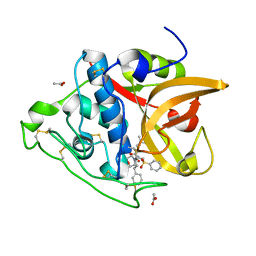 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with K11017 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-05-18 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
4O1G
 
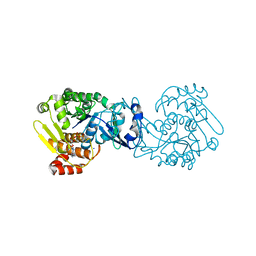 | | MTB adenosine kinase in complex with gamma-Thio-ATP | | Descriptor: | Adenosine kinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SODIUM ION | | Authors: | Dostal, J, Brynda, J, Hocek, M, Pichova, I. | | Deposit date: | 2013-12-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
4O1L
 
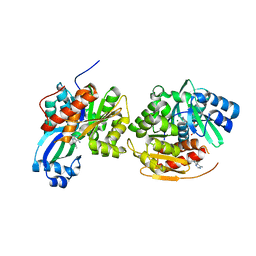 | | Human Adenosine Kinase in complex with inhibitor | | Descriptor: | 5-ethynyl-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Adenosine kinase, MAGNESIUM ION | | Authors: | Brynda, J, Dostal, J, Pichova, I, Hodcek, M. | | Deposit date: | 2013-12-16 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
1J9I
 
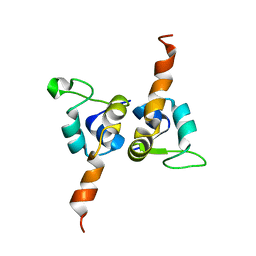 | | STRUCTURE OF THE DNA BINDING DOMAIN OF THE GPNU1 SUBUNIT OF LAMBDA TERMINASE | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | De Beer, T, Meyer, J, Ortega, M, Yang, Q, Maes, L, Duffy, C, Berton, N, Sippy, J, Overduin, M, Feiss, M, Catalano, C. | | Deposit date: | 2001-05-25 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into specific DNA recognition during the assembly of a viral genome packaging machine.
Mol.Cell, 9, 2002
|
|
3I8W
 
 | |
4PVV
 
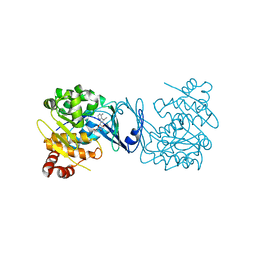 | | Micobacterial Adenosine Kinase in complex with inhibitor | | Descriptor: | 5-ethynyl-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Adenosine kinase | | Authors: | Pichova, I, Hocek, M, Dostal, J, Rezacova, P. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
3S3R
 
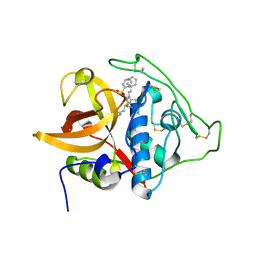 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with K11777 inhibitor | | Descriptor: | Cathepsin B-like peptidase (C01 family), Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2011-05-18 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Basis for Inhibition of Cathepsin B Drug Target from the Human Blood Fluke, Schistosoma mansoni.
J.Biol.Chem., 286, 2011
|
|
5OGR
 
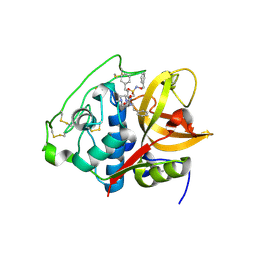 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with WRR286 inhibitor | | Descriptor: | 3-[[N-[4-METHYL-PIPERAZINYL]CARBONYL]-PHENYLALANINYL-AMINO]-5-PHENYL-PENTANE-1-SULFONIC ACID BENZYLOXY-AMIDE, ACETATE ION, Cathepsin B-like peptidase (C01 family) | | Authors: | Jilkova, A, Rezacova, P, Brynda, J, Mares, M. | | Deposit date: | 2017-07-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors.
Acs Infect Dis., 2020
|
|
5OGQ
 
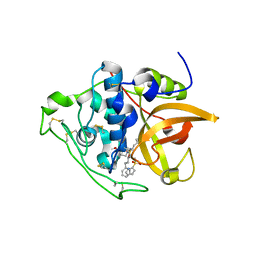 | | Structure of cathepsin B1 from Schistosoma mansoni in complex with WRR391 inhibitor | | Descriptor: | ACETATE ION, Cathepsin B-like peptidase (C01 family), ethyl 1-[[(2~{S})-3-(4-hydroxyphenyl)-1-oxidanylidene-1-[[(3~{S})-1-phenyl-5-pyridin-2-ylsulfonyl-pentan-3-yl]amino]propan-2-yl]carbamoyl]piperidine-4-carboxylate | | Authors: | Jilkova, A, Rezacova, P, Brynda, J, Mares, M. | | Deposit date: | 2017-07-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Druggable Hot Spots in the Schistosomiasis Cathepsin B1 Target Identified by Functional and Binding Mode Analysis of Potent Vinyl Sulfone Inhibitors.
Acs Infect Dis., 2020
|
|
8J00
 
 | | Human KCNQ2-CaM in complex with CBD | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, cannabidiol | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8IZY
 
 | | Human KCNQ2-CaM in complex with HN37 | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2, methyl N-[4-[(4-fluorophenyl)methyl-prop-2-ynyl-amino]-2,6-dimethyl-phenyl]carbamate | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J03
 
 | |
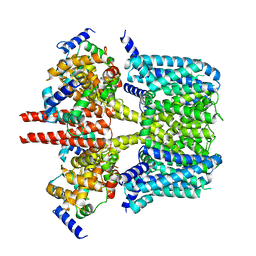
8J05
 
 | |
8J04
 
 | | Human KCNQ2-CaM-HN37 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, methyl N-[4-[(4-fluorophenyl)methyl-prop-2-ynyl-amino]-2,6-dimethyl-phenyl]carbamate | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J02
 
 | | Human KCNQ2(F104A)-CaM-PIP2-CBD complex in state II | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J01
 
 | | Human KCNQ2-CaM in complex with CBD and PIP2 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
6KBW
 
 | | Crystal structure of Tmm from Myroides profundi D25 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethylamine monooxygenase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Oxidation of trimethylamine to trimethylamine N-oxide facilitates high hydrostatic pressure tolerance in a generalist bacterial lineage.
Sci Adv, 7, 2021
|
|
4PW2
 
 | | Crystal structure of D-glucuronyl C5 epimerase | | Descriptor: | CITRIC ACID, D-glucuronyl C5 epimerase B | | Authors: | Ke, J, Qin, Y, Gu, X, Brunzelle, J.S, Xu, H.E, Ding, K. | | Deposit date: | 2014-03-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Study of d-Glucuronyl C5-epimerase.
J.Biol.Chem., 290, 2015
|
|
