2WZM
 
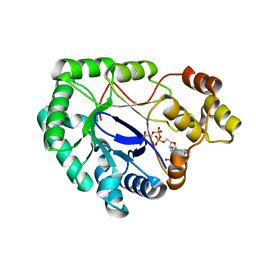 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
2VH0
 
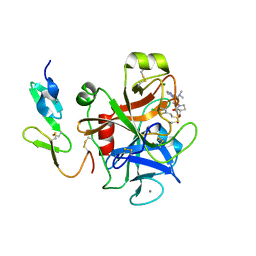 | | Structure and property based design of factor Xa inhibitors:biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-[(3S)-1-(4-{2-[(dimethylamino)methyl]-1H-imidazol-1-yl}-2-fluorophenyl)-2-oxopyrrolidin-3-yl]ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Convery, M.A, Diallo, H, Hortense, E, Irving, W.R, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and property based design of factor Xa inhibitors: biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs.
Bioorg. Med. Chem. Lett., 18, 2008
|
|
2VS8
 
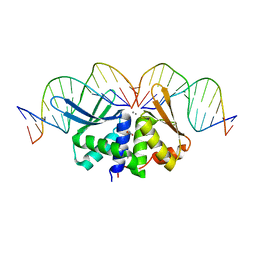 | | The crystal structure of I-DmoI in complex with DNA and Mn | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP* GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP *CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP *GP*TP*AP*A)-3', ... | | Authors: | Marcaida, M.J, Prieto, J, Redondo, P, Nadra, A.D, Alibes, A, Serrano, L, Grizot, S, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2008-04-21 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VWE
 
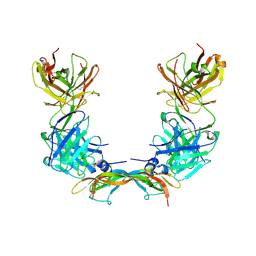 | | Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralizing Antibody Fab Fragment | | Descriptor: | ANTI-VEGF-B MONOCLONAL ANTIBODY, VASCULAR ENDOTHELIAL GROWTH FACTOR B | | Authors: | Leonard, P, Scotney, P.D, Jabeen, T, Iyer, S, Fabri, L.J, Nash, A.D, Acharya, K.R. | | Deposit date: | 2008-06-23 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Vascular Endothelial Growth Factor-B in Complex with a Neutralising Antibody Fab Fragment.
J.Mol.Biol., 384, 2008
|
|
5CSE
 
 | | Streptavidin-S112Y-K121E Complexed with Palladium-Containing Biotin Ligand | | Descriptor: | CHLORIDE ION, Streptavidin, chloro{di-tert-butyl[2-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)ethyl]-lambda~5~-phosphanyl}(1-phenylprop-1-ene-1,3-diyl-kappa~2~C~1~,C~3~)palladium | | Authors: | Finke, A.D, Vera, L, Marsh, M, Chatterjee, A, Ward, T.R. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | An enantioselective artificial Suzukiase based on the biotin-streptavidin technology.
Chem Sci, 7, 2016
|
|
1L5Y
 
 | | CRYSTAL STRUCTURE OF MG2+ / BEF3-BOUND RECEIVER DOMAIN OF SINORHIZOBIUM MELILOTI DCTD | | Descriptor: | BERYLLIUM DIFLUORIDE, BERYLLIUM TETRAFLUORIDE ION, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Park, S, Meyer, M, Jones, A.D, Yennawar, H.P, Yennawar, N.H, Nixon, B.T. | | Deposit date: | 2002-03-08 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two-component signaling in the AAA + ATPase DctD: binding Mg2+ and BeF3- selects between alternate dimeric states of the receiver domain
FASEB J., 16, 2002
|
|
5D5S
 
 | | Crystal structure of methionine gamma-lyase from Citrobacter freundii, S339A mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Anufrieva, N.V, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2015-08-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of methionine gamma-lyase from Citrobacter freundii, S339A mutant
To Be Published
|
|
2XUT
 
 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
5BSX
 
 | |
1KUQ
 
 | | CRYSTAL STRUCTURE OF T3C MUTANT S15 RIBOSOMAL PROTEIN IN COMPLEX WITH 16S RRNA | | Descriptor: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, SULFATE ION | | Authors: | Nikulin, A.D, Tishchenko, S, Revtovich, S, Ehresmann, B, Ehresmann, C, Dumas, P, Garber, M, Nikonov, S, Nevskaya, N. | | Deposit date: | 2002-01-22 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Role of N-terminal helix in interaction of ribosomal protein S15 with 16S rRNA.
Biochemistry Mosc., 69, 2004
|
|
2WYJ
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1S)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2WZT
 
 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
5BSY
 
 | |
1MNB
 
 | | BIV TAT PEPTIDE (RESIDUES 68-81), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BIV TAR RNA, BIV TAT PEPTIDE | | Authors: | Puglisi, J.D, Chen, L, Blanchard, S, Frankel, A.D. | | Deposit date: | 1996-07-25 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex.
Science, 270, 1995
|
|
2VH6
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with biaryl P4 motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-{(3S)-1-[3-fluoro-2'-(methylsulfonyl)biphenyl-4-yl]-2-oxopyrrolidin-3-yl}ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Chung, C.W, Convery, M.A, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A.N, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E. | | Deposit date: | 2007-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Biaryl P4 Motifs.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5DBV
 
 | | Structure of a C269A mutant of propionaldehyde dehydrogenase from the Clostridium phytofermentans fucose utilisation bacterial microcompartment | | Descriptor: | ACETATE ION, Aldehyde Dehydrogenase, COENZYME A, ... | | Authors: | Tuck, L.R, Altenbach, K, Ang, T.F, Crawshaw, A.D, Campopiano, D.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2015-08-22 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Insight into Coenzyme A cofactor binding and the mechanism of acyl-transfer in an acylating aldehyde dehydrogenase from Clostridium phytofermentans.
Sci Rep, 6, 2016
|
|
1N2W
 
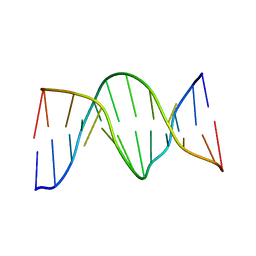 | | Solution Structure of 8OG:G mismatch containing duplex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(8OG)P*GP*CP*G)-3' | | Authors: | Thiviyanathan, V, Somasunderam, A.D, Hazra, T.K, Mitra, S, Gorenstein, D.G. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing 8-Hydroxy-2'-Deoxyguanosine Opposite Deoxyguanosine
J.Mol.Biol., 325, 2003
|
|
2Y37
 
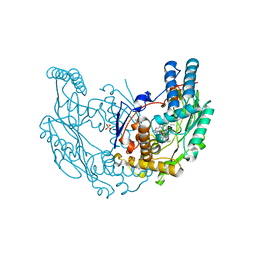 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 2-[(1R)-3-amino-1-phenyl-propoxy]-4-chloro-benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T.N, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2010-12-19 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2XRU
 
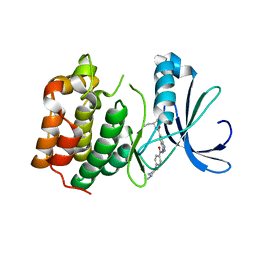 | | AURORA-A T288E COMPLEXED WITH PHA-828300 | | Descriptor: | 3-({[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]CARBONYL}AMINO)-N-[(1R)-1-PHENYLPROPYL]-1H-THIENO[3,2-C]PYRAZOLE-5-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Bindi, S, Fancelli, D, Alli, C, Berta, D, Bertrand, J.A, Cameron, A.D, Cappella, P, Carpinelli, P, Cervi, G, Croci, W, D'Anello, M, Forte, B, LauraGiorgini, M, Marsiglio, A, Moll, J, Pesenti, E, Pittala, V, Pulici, M, Riccardi-Sirtori, F, Roletto, F, Soncini, C, Storici, P, Varasi, M, Volpi, D, Zugnoni, P, Vianello, P. | | Deposit date: | 2010-09-22 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thieno[3,2-C]Pyrazoles: A Novel Class of Aurora Inhibitors with Favorable Antitumor Activity.
Bioorg.Med.Chem., 18, 2010
|
|
2XVM
 
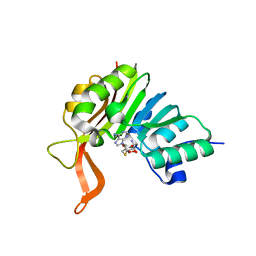 | | Crystal structure of the tellurite detoxification protein TehB from E. coli in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TELLURITE RESISTANCE PROTEIN TEHB | | Authors: | Choudhury, H.G, Cameron, A.D, Iwata, S, Beis, K. | | Deposit date: | 2010-10-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure and Mechanism of the Chalcogen Detoxifying Protein Tehb from Escherichia Coli.
Biochem.J., 435, 2011
|
|
1KER
 
 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
2VS7
 
 | | The crystal structure of I-DmoI in complex with DNA and Ca | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP *CP*TP*TP*AP*CP*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP *GP*TP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*G)-3', ACETATE ION, ... | | Authors: | Marcaida, M.J, Prieto, J, Redondo, P, Nadra, A.D, Alibes, A, Serrano, L, Grizot, S, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2008-04-21 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of I-Dmoi in Complex with its Target DNA Provides New Insights Into Meganuclease Engineering.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1N6Q
 
 | | HIV-1 Reverse Transcriptase Crosslinked to pre-translocation AZTMP-terminated DNA (complex N) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3', 5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, I, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-11 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 Reverse Transcriptase with Pre- and Post-translocation AZTMP-terminated DNA
Embo J., 21, 2002
|
|
1N5Y
 
 | | HIV-1 Reverse Transcriptase Crosslinked to Post-Translocation AZTMP-Terminated DNA (Complex P) | | Descriptor: | 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3', 5'-D(*AP*TP*GP*C*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Sarafianos, S.G, Clark Jr, A.D, Das, K, Tuske, S, Birktoft, J.J, Ilankumaran, P, Ramesha, A.R, Sayer, J.M, Jerina, D.M, Boyer, P.L, Hughes, S.H, Arnold, E. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of HIV-1 Reverse Transcriptase with Pre-Translocation and Post-Translocation AZTMP-Terminated DNA
Embo J., 21, 2002
|
|
3HCT
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the P1 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
