4NJ9
 
 | | Crystal structure of Fab 8B10 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, 8B10 heavy chain, 8B10 light chain, ... | | Authors: | Stanfield, R.L, Romesberg, F.E, Zimmermann, J, Wilson, I.A. | | Deposit date: | 2013-11-08 | | Release date: | 2014-11-12 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptive Mutations Alter Antibody Structure and Dynamics during Affinity Maturation.
Biochemistry, 54, 2015
|
|
4NJA
 
 | | Crystal structure of Fab 6C8 in complex with MPTS | | Descriptor: | 6C8 heavy chain, 6C8 light chain, 8-methoxypyrene-1,3,6-trisulfonic acid, ... | | Authors: | Stanfield, R.L, Romesberg, F.E, Zimmermann, J, Wilson, I.A. | | Deposit date: | 2013-11-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Adaptive Mutations Alter Antibody Structure and Dynamics during Affinity Maturation.
Biochemistry, 54, 2015
|
|
7EAN
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 6D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 cross-neutralizing mAb 6D6, Light chain of SARS-CoV-2 cross-neutralizing mAb 6D6, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
7EAM
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 7D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
6CKV
 
 | |
5YA5
 
 | | CRYSTAL STRUCTURE OF c-MET IN COMPLEX WITH NOVEL INHIBITOR | | Descriptor: | 2-[3-(4-methoxybenzyl)[1,2,4]triazolo[3,4-b][1,3,4]thiadiazol-6-yl]-1H-indole, Hepatocyte growth factor receptor | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-08-30 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery, optimization and biological evaluation for novel c-Met kinase inhibitors
Eur J Med Chem, 143, 2018
|
|
4GG5
 
 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 3-(4-methylpiperazin-1-yl)-N-(3-nitrobenzyl)-7-(trifluoromethyl)quinolin-5-amine, Hepatocyte growth factor receptor | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4GG7
 
 | | Crystal structure of cMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-nitrobenzyl)-6-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-2-(trifluoromethyl)pyrido[2,3-d]pyrimidin-4-amine | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8TU5
 
 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 27 | | Descriptor: | 1-[(1S,5S,6S)-6-methyl-6-{[(6M,8R)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}-2-azabicyclo[3.2.0]heptan-2-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
8TU3
 
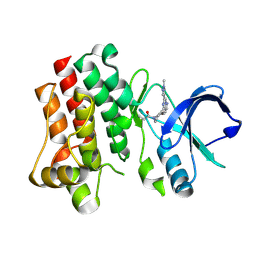 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 10 | | Descriptor: | 1-[(4R)-4-{[(6P,8S)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}azepan-1-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
8TU4
 
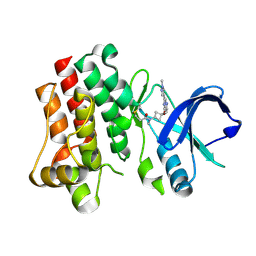 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 25 | | Descriptor: | N-methyl-N-[(1S,3r)-3-methyl-3-{[(6M,8S)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}cyclobutyl]propanamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
8VZN
 
 | |
8VZO
 
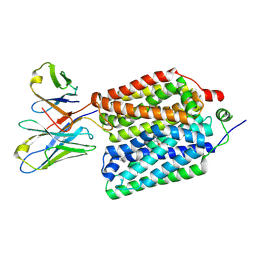 | |
7DPF
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion | | Descriptor: | Capsid protein VP4, PALMITIC ACID, VP2, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
6KI2
 
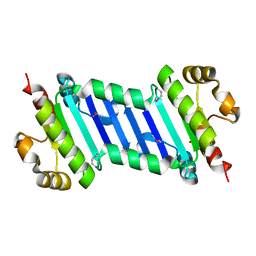 | |
6KI1
 
 | |
6DDI
 
 | | Crystal Structure of the human BRD2 BD1 bromodomain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
6OHR
 
 | |
6OHO
 
 | | Structure of human Phospholipase D2 catalytic domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Phospholipase D2, ... | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2019-04-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human PLD structures enable drug design and characterization of isoenzyme selectivity.
Nat.Chem.Biol., 16, 2020
|
|
6OHP
 
 | |
6OHQ
 
 | |
6DDJ
 
 | | Crystal Structure of the human BRD2 BD2 bromodimain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
6DUK
 
 | | EGFR with an allosteric inhibitor | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-{1-oxo-6-[4-(piperazin-1-yl)phenyl]-1,3-dihydro-2H-isoindol-2-yl}-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Park, E, Eck, M.J. | | Deposit date: | 2018-06-21 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single and Dual Targeting of Mutant EGFR with an Allosteric Inhibitor.
Cancer Discov, 9, 2019
|
|
5D41
 
 | | EGFR kinase domain in complex with mutant selective allosteric inhibitor | | Descriptor: | (2R)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-2-phenyl-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Yun, C.-H, Park, E, Eck, M.J. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors.
Nature, 534, 2016
|
|
7UAP
 
 | |
