7XTC
 
 | | Serotonin 7 (5-HT7) receptor-Gs-Nb35 complex | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT8
 
 | | Serotonin 4 (5-HT4) receptor-Gs-Nb35 complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7XT9
 
 | | Serotonin 4 (5-HT4) receptor-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7DFG
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir | | Descriptor: | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Li, Z, Zhou, Z, Yu, X. | | Deposit date: | 2020-11-08 | | Release date: | 2021-11-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for repurpose and design of nucleotide drugs for treating COVID-19
To Be Published
|
|
7DFH
 
 | |
7DOI
 
 | |
7DOK
 
 | |
7CMU
 
 | | Dopamine Receptor D3R-Gi-Pramipexole complex | | Descriptor: | (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7CMV
 
 | | Dopamine Receptor D3R-Gi-PD128907 complex | | Descriptor: | (4aR,10bR)-4-propyl-3,4a,5,10b-tetrahydro-2H-chromeno[4,3-b][1,4]oxazin-9-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | Descriptor: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8JRN
 
 | | Structure of E6AP-E6 complex in Att1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRQ
 
 | | Structure of E6AP-E6 complex in Det1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRP
 
 | | Structure of E6AP-E6 complex in Att3 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRR
 
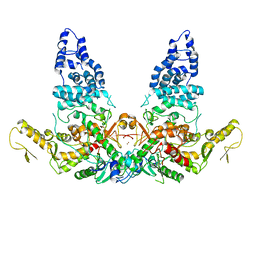 | | Structure of E6AP-E6 complex in Det2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRO
 
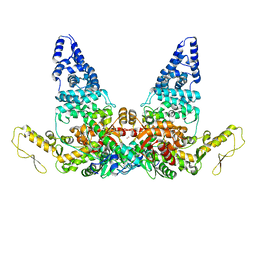 | | Structure of E6AP-E6 complex in Att2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
7BQT
 
 | | Epstein-Barr virus, C12 portal dodecamer | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BQX
 
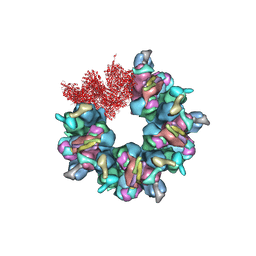 | | Epstein-Barr virus, C5 portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BR8
 
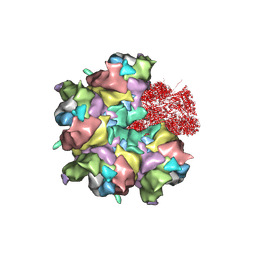 | | Epstein-Barr virus, C5 penton vertex, CATC absent. | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BSI
 
 | |
7BR7
 
 | | Epstein-Barr virus, C1 portal-proximal penton vertex, CATC binding | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7DFB
 
 | | Crystal of Arrestin2-V2Rpp-6-7-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DF9
 
 | | Crystal of Arrestin2-V2Rpp-1-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFA
 
 | | Crystal of Arrestin2-V2Rpp-4-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFC
 
 | | Crystal of Arrestin2-V2Rpp-3-Fab30 complex | | Descriptor: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | Authors: | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
