4UA8
 
 | |
4UAC
 
 | | EUR_01830 with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Carbohydrate ABC transporter substrate-binding protein, ... | | Authors: | Koropatkin, N.M, Orlovsky, N.I. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale.
Mol.Microbiol., 95, 2015
|
|
7O7B
 
 | |
1P1W
 
 | |
1P1Q
 
 | |
8EHX
 
 | | cryo-EM structure of TMEM63B in LMNG | | Descriptor: | CSC1-like protein 2 | | Authors: | Zheng, W, Fu, T.M, Holt, J.R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | TMEM63 proteins function as monomeric high-threshold mechanosensitive ion channels.
Neuron, 111, 2023
|
|
8EHW
 
 | |
6WHG
 
 | | PI3P and calcium bound full-length TRPY1 in detergent | | Descriptor: | (2R)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1S,2S,3S,4S,5S,6R)-2,3,4,6-tetrahydroxy-5-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl hexadecanoate, CALCIUM ION, Calcium channel YVC1 | | Authors: | Ahmed, T, Moiseenkova-Bell, V.Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the ancient TRPY1 channel from Saccharomyces cerevisiae reveals mechanisms of modulation by lipids and calcium.
Structure, 30, 2022
|
|
6XV7
 
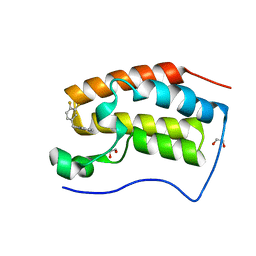 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 2 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[3,4-bis(fluoranyl)phenyl]methyl]-~{N},3-dimethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XUZ
 
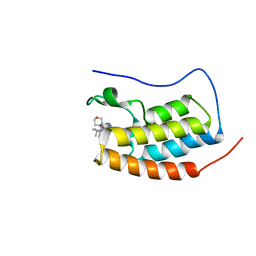 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 4 | | Descriptor: | 6-[1-[(2~{S})-1-methoxypropan-2-yl]-6-[(3~{S})-3-methylmorpholin-4-yl]imidazo[4,5-c]pyridin-2-yl]-3-methyl-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XV3
 
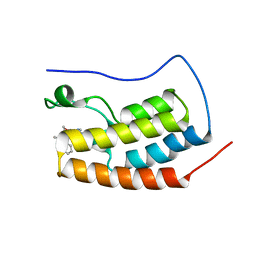 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 3 | | Descriptor: | 3-methyl-6-[6-[(3~{S})-3-methylmorpholin-4-yl]-1-[(1~{S})-1-phenylethyl]imidazo[4,5-c]pyridin-2-yl]-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8R07
 
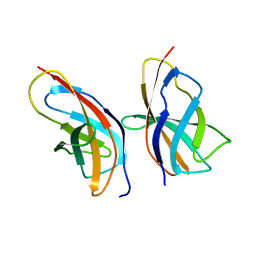 | | C-terminal Rel-homology Domain of NFAT1 | | Descriptor: | Nuclear factor of activated T-cells, cytoplasmic 2 | | Authors: | Zak, K.M, Boettcher, J. | | Deposit date: | 2023-10-30 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Ligandability assessment of the C-terminal Rel-homology domain of NFAT1.
Arch Pharm, 357, 2024
|
|
8R3F
 
 | | C-terminal Rel-homology Domain of NFAT1 | | Descriptor: | (4~{S})-6-fluoranyl-3,4-dihydro-2~{H}-chromen-4-amine, Nuclear factor of activated T-cells, cytoplasmic 2 | | Authors: | Zak, K.M, Boettcher, J. | | Deposit date: | 2023-11-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ligandability assessment of the C-terminal Rel-homology domain of NFAT1.
Arch Pharm, 357, 2024
|
|
6XVC
 
 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 1 | | Descriptor: | (4~{R})-4-[(1~{R})-1-[7-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)quinolin-5-yl]oxyethyl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D, Wolkerstorfer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.098 Å) | | Cite: | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8P0F
 
 | | Crystal structure of the VCB complex with compound 1. | | Descriptor: | (3~{R},5~{R})-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-5-oxidanyl-2-oxidanylidene-1-pyridin-2-yl-piperidine-3-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Drugit: Crowd-sourcing molecular design of non-peptidic VHL binders
Chemrxiv, 2023
|
|
4E9O
 
 | | Vaccinia D8L ectodomain structure | | Descriptor: | IMV membrane protein, IODIDE ION | | Authors: | Matho, M.H, Zajonc, D.M. | | Deposit date: | 2012-03-21 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and Biochemical Characterization of the Vaccinia Virus Envelope Protein D8 and Its Recognition by the Antibody LA5.
J.Virol., 86, 2012
|
|
4ETQ
 
 | |
4DX1
 
 | | Crystal structure of the human TRPV4 ankyrin repeat domain | | Descriptor: | GLYCEROL, PHOSPHATE ION, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Inada, H, Gaudet, R. | | Deposit date: | 2012-02-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel.
Biochemistry, 51, 2012
|
|
4DX2
 
 | | Crystal structure of the human TRPV4 ankyrin repeat domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Transient receptor potential cation channel subfamily V member 4, ... | | Authors: | Inada, H, Gaudet, R. | | Deposit date: | 2012-02-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel.
Biochemistry, 51, 2012
|
|
7KI6
 
 | |
7KI4
 
 | |
4EBQ
 
 | |
6QUU
 
 | | Crystal Structure of KRAS-G12D in complex with GMP-PCP | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Fischer, G, Kessler, D, Muellauer, B, Wolkerstorfer, B. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | KRAS Binders Hidden in Nature.
Chemistry, 25, 2019
|
|
6QUW
 
 | | Crystal Structure of KRAS-G12D in Complex with Natural Product-Like Compound 9b | | Descriptor: | (3~{a}~{R},8~{b}~{S})-2,2,3~{a},8~{b}-tetramethyl-3,4-dihydro-1~{H}-pyrrolo[2,3-b]indole, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Fischer, G, Kessler, D, Muellauer, B, Wolkerstorfer, B. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | KRAS Binders Hidden in Nature.
Chemistry, 25, 2019
|
|
6QUV
 
 | | Crystal Structure of KRAS-G12D in complex with GMP-PCP and compound 15R | | Descriptor: | (6~{a}~{R},11~{b}~{S})-6~{a}-(1,4-dimethylpiperidin-4-yl)-7,11~{b}-dihydro-6~{H}-indolo[2,3-c]isoquinolin-5-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Fischer, G, Kessler, D, Muellauer, B, Wolkerstorfer, B. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | KRAS Binders Hidden in Nature.
Chemistry, 25, 2019
|
|
