3JVL
 
 | | Crystal structure of bromodomain 2 of mouse Brd4 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromodomain-containing protein 4 | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
3JZ9
 
 | | Crystal structure of the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
3HXE
 
 | | Engineered RabGGTase in complex with a peptidomimetic inhibitor (compound 37) | | Descriptor: | CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, Geranylgeranyl transferase type-2 subunit beta, ... | | Authors: | Guo, Z, Alexandrov, K, Waldmann, H, Goody, R.S, Blankenfeldt, W. | | Deposit date: | 2009-06-20 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, and characterization of Peptide-based rab geranylgeranyl transferase inhibitors
J.Med.Chem., 52, 2009
|
|
3JVK
 
 | | Crystal structure of bromodomain 1 of mouse Brd4 in complex with histone H3-K(ac)14 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, histone H3.3 peptide | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
3KH3
 
 | |
3KH4
 
 | |
3PZ4
 
 | | Crystal structure of FTase(ALPHA-subunit; BETA-subunit DELTA C10) in complex with BMS3 and lipid substrate FPP | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, FARNESYL DIPHOSPHATE, Protein farnesyltransferase subunit beta, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ3
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS-analogue 14 | | Descriptor: | 4-({(3R)-7-(5-formylfuran-2-yl)-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl benzylcarbamate, CALCIUM ION, Geranylgeranyl transferase type-2 subunit alpha, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ1
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS3 | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3PZ2
 
 | | Crystal structure of RabGGTase(DELTA LRR; DELTA IG) in Complex with BMS3 and lipid substrate GGPP | | Descriptor: | (3R)-3-benzyl-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepine-7-carbonitrile, CALCIUM ION, GERANYLGERANYL DIPHOSPHATE, ... | | Authors: | Guo, Z, Bon, R.S, Stigter, E.A, Waldmann, H, Alexandrov, K, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2010-12-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Guided Development of Selective RabGGTase Inhibitors.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3R76
 
 | | Crystal structure of 2-amino-2-desoxyisochorismate synthase (ADIC) synthase PhzE from Burkholderia lata 383 in complex with benzoate, pyruvate and glutamine | | Descriptor: | Anthranilate/para-aminobenzoate synthases component I, BENZOIC ACID, MAGNESIUM ION, ... | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
3REB
 
 | |
3R74
 
 | | Crystal structure of 2-amino-2-desoxyisochorismate synthase (ADIC) synthase PhzE from Burkholderia lata 383 | | Descriptor: | Anthranilate/para-aminobenzoate synthases component I | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
3S9G
 
 | |
3R75
 
 | | Crystal structure of 2-amino-2-desoxyisochorismate synthase (ADIC) synthase PhzE from Burkholderia lata 383 in complex with benzoate, pyruvate, glutamine and contaminating Zn2+ | | Descriptor: | Anthranilate/para-aminobenzoate synthases component I, BENZOIC ACID, MAGNESIUM ION, ... | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
3REA
 
 | |
3R77
 
 | | Crystal structure of the D38A mutant of isochorismatase PhzD from Pseudomonas fluorescens 2-79 in complex with 2-amino-2-desoxyisochorismate ADIC | | Descriptor: | (5S,6S)-6-amino-5-[(1-carboxyethenyl)oxy]cyclohexa-1,3-diene-1-carboxylic acid, CHLORIDE ION, Probable isochorismatase | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
3QBT
 
 | | Crystal structure of OCRL1 540-678 in complex with Rab8a:GppNHp | | Descriptor: | Inositol polyphosphate 5-phosphatase OCRL-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hou, X, Hagemann, N, Schoebel, S, Blankenfeldt, W, Goody, R.S, Erdmann, K.S, Itzen, A. | | Deposit date: | 2011-01-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1.
Embo J., 30, 2011
|
|
1SDJ
 
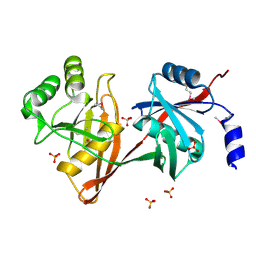 | | X-RAY STRUCTURE OF YDDE_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ET25. | | Descriptor: | Hypothetical protein yddE, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Skarina, T, Korniyenko, Y, Savchenko, A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6RMS
 
 | |
1O7I
 
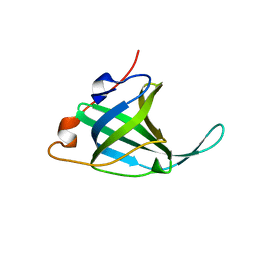 | |
4AY7
 
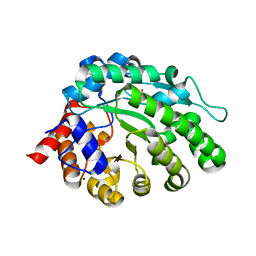 | | methyltransferase from Methanosarcina mazei | | Descriptor: | MAGNESIUM ION, METHYLCOBALAMIN: COENZYME M METHYLTRANSFERASE, ZINC ION | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeld, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5G3U
 
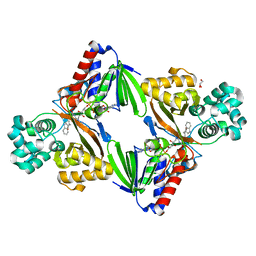 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum in complex with its inhibitor 2-(1H-indol-3-ylmethyl)prop-2- enoic acid | | Descriptor: | 2-[(1H-indol-3-yl)methyl]prop-2-enoic acid, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Biosynthesis of Violacein, Structure and Function of l-Tryptophan Oxidase VioA from Chromobacterium violaceum.
J.Biol.Chem., 291, 2016
|
|
5G3S
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum - Samarium derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Biosynthesis of Violacein: Structure and Function of L-Tryptophan Oxidase Vioa Chromobacterium Violaceum
J.Biol.Chem., 291, 2016
|
|
5G3T
 
 | | The structure of the L-tryptophan oxidase VioA from Chromobacterium violaceum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Krausze, J, Rabe, J, Moser, J. | | Deposit date: | 2016-05-01 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biosynthesis of Violacein: Structure and Function of L-Tryptophan Oxidase Vioa Chromobacterium Violaceum
J.Biol.Chem., 291, 2016
|
|
