3AQA
 
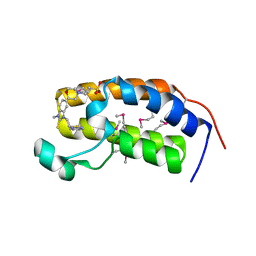 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
2DVS
 
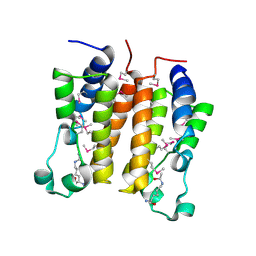 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVR
 
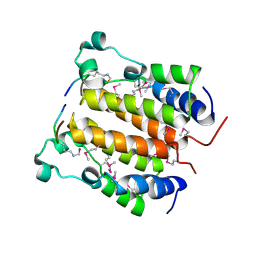 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVQ
 
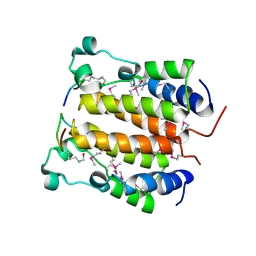 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | Bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
7X73
 
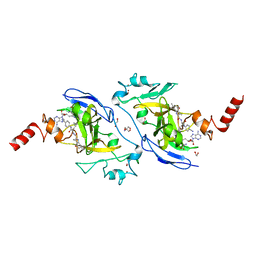 | | Structure of G9a in complex with RK-701 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nat Commun, 14, 2023
|
|
6M4W
 
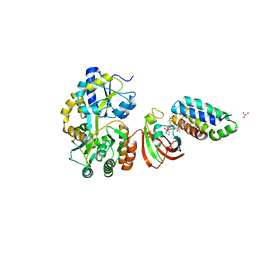 | | Crystal structure of MBP fused split FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
5WRM
 
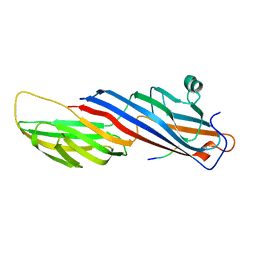 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y658 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
7XUD
 
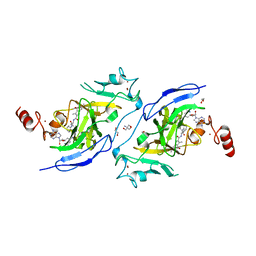 | | Structure of G9a in complex with compound 26a | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUA
 
 | | Structure of G9a in complex with compound 10a | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUC
 
 | | Structure of G9a in complex with compound 11a | | Descriptor: | 1,2-ETHANEDIOL, 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-phenylazanyl-hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUB
 
 | | Structure of G9a in complex with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
5X17
 
 | | Crystal structure of murine CK1d in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, SULFATE ION | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
5WRL
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y628 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.095 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
5WRK
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y608 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1, NICKEL (II) ION | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
6M4V
 
 | | Crystal structure of MBP fused split FKBP in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
6M4U
 
 | | Crystal structure of FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | CHLORIDE ION, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
7VQT
 
 | | Crystal structure of LSD1 in complex with compound 5 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-4-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQS
 
 | | Crystal structure of LSD1 in complex with compound 4 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQU
 
 | | Crystal structure of LSD1 in complex with compound S1427 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7W3L
 
 | | Crystal structure of LSD1 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE1
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE2
 
 | | Crystal structure of LSD2 in complex with trans-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE3
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, CITRATE ANION, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
5H34
 
 | | Crystal structure of the C-terminal domain of methionyl-tRNA synthetase (MetRS-C) in Nanoarchaeum equitans | | Descriptor: | Methionine-tRNA ligase | | Authors: | Suzuki, H, Kaneko, A, Yamamoto, T, Nambo, M, Umehara, T, Yoshida, H, Park, S.Y, Tamura, K. | | Deposit date: | 2016-10-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Binding Properties of Split tRNA to the C-terminal Domain of Methionyl-tRNA Synthetase of Nanoarchaeum equitans.
J. Mol. Evol., 84, 2017
|
|
3VUZ
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with AAM-1 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](hexyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
