6AMB
 
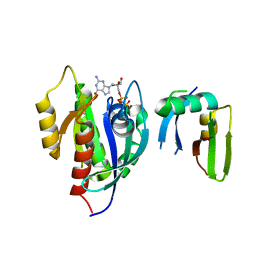 | | Crystal Structure of the Afadin RA1 domain in complex with HRAS | | Descriptor: | Afadin, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Smith, M.J, Ishiyama, N, Ikura, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of AF6-RAS association and its implications in mixed-lineage leukemia.
Nat Commun, 8, 2017
|
|
6TGC
 
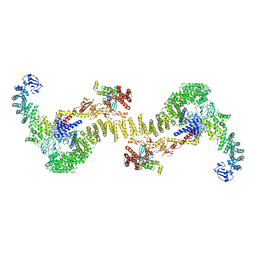 | | CryoEM structure of the ternary DOCK2-ELMO1-RAC1 complex. | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6TGB
 
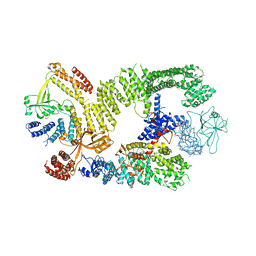 | | CryoEM structure of the binary DOCK2-ELMO1 complex | | Descriptor: | Dedicator of cytokinesis protein 2, Engulfment and cell motility protein 1 | | Authors: | Chang, L, Yang, J, Chang, J.H, Zhang, Z, Boland, A, McLaughlin, S.H, Abu-Thuraia, A, Killoran, R.C, Smith, M.J, Cote, J.F, Barford, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
2AZR
 
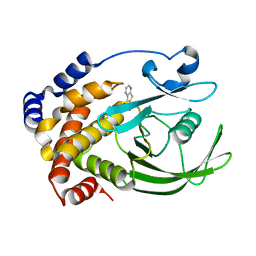 | | Crystal structure of PTP1B with Bicyclic Thiophene inhibitor | | Descriptor: | 3-(CARBOXYMETHOXY)THIENO[2,3-B]PYRIDINE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
2B07
 
 | | Crystal structure of PTP1B with Tricyclic Thiophene inhibitor. | | Descriptor: | 6-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}-3-(CARBOXYMETHOXY)THIENO[3,2-B][1]BENZOTHIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase, non-receptor type 1 | | Authors: | Moretto, A.F, Kirincich, S.J, Xu, W.X, Smith, M.J, Wan, Z.K, Wilson, D.P, Follows, B.C, Binnun, E, Joseph-McCarthy, D, Foreman, K, Erbe, D.V, Zhang, Y.L, Tam, S.K, Tam, S.Y, Lee, J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bicyclic and tricyclic thiophenes as protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem., 14, 2006
|
|
6UKA
 
 | | Crystal structure of RHOG and ELMO complex | | Descriptor: | Engulfment and cell motility protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jo, C.H, Killoran, R.C, Smith, M.J. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
3L6Y
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3L6X
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin, SULFATE ION | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
2KIS
 
 | | Solution structure of CA150 FF1 domain and FF1-FF2 interdomain linker | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Murphy, J.M, Hansen, D, Wiesner, S, Muhandiram, D, Borg, M, Smith, M.J, Sicheri, F, Kay, L.E, Forman-Kay, J.D, Pawson, T. | | Deposit date: | 2009-05-08 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural studies of FF domains of the transcription factor CA150 provide insights into the organization of FF domain tandem arrays.
J.Mol.Biol., 393, 2009
|
|
1QC8
 
 | | NMR STRUCTURE OF TAU EXON 10 SPLICING REGULATORY ELEMENT RNA | | Descriptor: | TAU EXON 10 SPLICING REGULATORY ELEMENT RNA | | Authors: | Varani, L, Spillantini, M.G, Klug, A, Goedert, M, Varani, G. | | Deposit date: | 1999-05-18 | | Release date: | 1999-08-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2QBR
 
 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 5-[3-(BENZYLAMINO)PHENYL]-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBQ
 
 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-{3-[(3,3,5,5-TETRAMETHYLCYCLOHEXYL)AMINO]PHENYL}THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBP
 
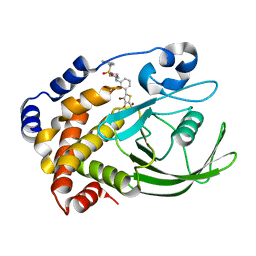 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2MSD
 
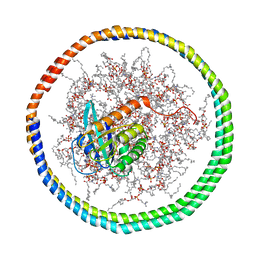 | | NMR data-driven model of GTPase KRas-GNP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSC
 
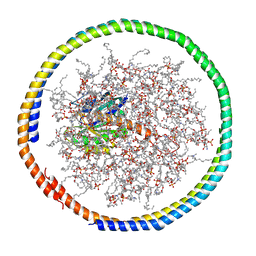 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2VXS
 
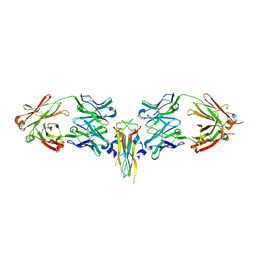 | | Structure of IL-17A in complex with a potent, fully human neutralising antibody | | Descriptor: | FAB FRAGMENT, INTERLEUKIN-17A, SULFATE ION | | Authors: | Gerhardt, S, Hargreaves, D, Pauptit, R.A, Davies, R.A, Russell, C, Welsh, F, Tuske, S.J, Coales, S.J, Hamuro, Y, Needham, M.R.C, Langham, C, Barker, W, Bell, P, Aziz, A, Smith, M.J, Dawson, S, Abbott, W.M. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of Il-17A in Complex with a Potent, Fully Human Neutralising Antibody.
J.Mol.Biol., 394, 2009
|
|
2QBS
 
 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-[3-(CYCLOHEXYLAMINO)PHENYL]THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
