6RMH
 
 | | The Rigid-body refined model of the normal Huntingtin. | | Descriptor: | Huntingtin | | Authors: | Jung, T, Tamo, G, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2019-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
6YEJ
 
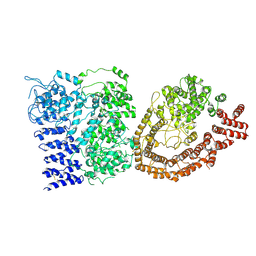 | | Cryo-EM structure of the Full-length disease type human Huntingtin | | Descriptor: | Huntingtin | | Authors: | Tame, G, Jung, T, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2020-03-24 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (18.200001 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
7XMH
 
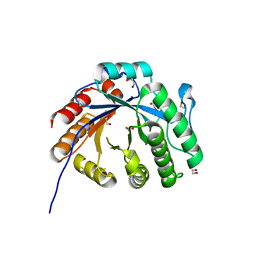 | | Crystal structure of a rice class IIIb chitinase, Oschib2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative class III chitinase | | Authors: | Jun, T, Tomoya, T, Tomoyuki, N, Takayuki, O. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Characterization of two rice GH18 chitinases belonging to family 8 of plant pathogenesis-related proteins.
Plant Sci., 326, 2023
|
|
4UW2
 
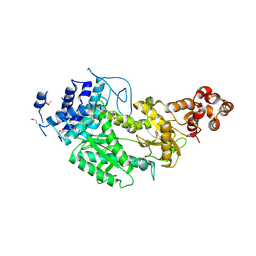 | | Crystal structure of Csm1 in T.onnurineus | | Descriptor: | CSM1 | | Authors: | Jung, T.Y, An, Y, Park, K.H, Lee, M.H, Oh, B.H, Woo, E.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-03-25 | | Last modified: | 2015-09-23 | | Method: | X-RAY DIFFRACTION (2.632 Å) | | Cite: | Crystal Structure of the Csm1 Subunit of the Csm Complex and its Single-Stranded DNA-Specific Nuclease Activity.
Structure, 23, 2015
|
|
5G4D
 
 | |
7R54
 
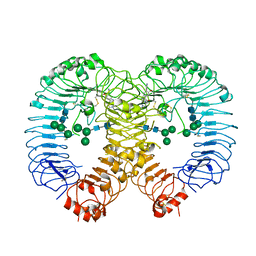 | | Crystal structure of human TLR8 in complex with Compound 4 | | Descriptor: | (5-methoxy-6-pyridin-4-yl-1~{H}-indazol-3-yl)-(4-methylpiperazin-1-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.836 Å) | | Cite: | Structure-Based Optimization of a Fragment-like TLR8 Binding Screening Hit to an In Vivo Efficacious TLR7/8 Antagonist.
Acs Med.Chem.Lett., 13, 2022
|
|
7R53
 
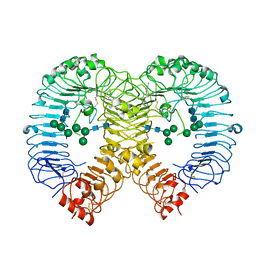 | | Crystal structure of human TLR8 in complex with Compound 15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-cyclopropyl-6-(2,6-dimethylpyridin-4-yl)-~{N}-[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]-1~{H}-indazol-3-amine, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.121 Å) | | Cite: | Structure-Based Optimization of a Fragment-like TLR8 Binding Screening Hit to an In Vivo Efficacious TLR7/8 Antagonist.
Acs Med.Chem.Lett., 13, 2022
|
|
7R52
 
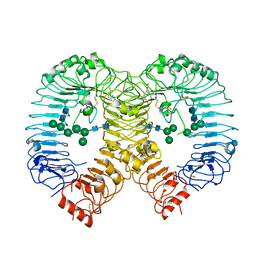 | | Crystal structure of human TLR8 in complex with Compound 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methoxy-6-pyridin-4-yl-1~{H}-indole, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.943 Å) | | Cite: | Structure-Based Optimization of a Fragment-like TLR8 Binding Screening Hit to an In Vivo Efficacious TLR7/8 Antagonist.
Acs Med.Chem.Lett., 13, 2022
|
|
6TY5
 
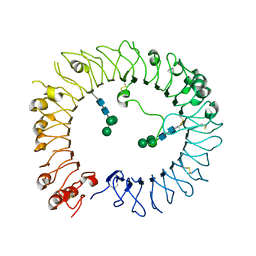 | | Crystal structure of human TLR8 in complex with Compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-7-(7-methyl-2-piperidin-4-yl-indazol-5-yl)furo[3,2-c]pyridin-4-one, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Target-Based Identification and Optimization of 5-Indazol-5-yl Pyridones as Toll-like Receptor 7 and 8 Antagonists Using a Biochemical TLR8 Antagonist Competition Assay.
J.Med.Chem., 63, 2020
|
|
8PFI
 
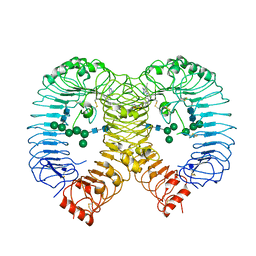 | | Crystal structure of human TLR8 in complex with compound 34 | | Descriptor: | (3~{S})-~{N}-[4-[[5-(1,6-dimethylpyrazolo[3,4-b]pyridin-4-yl)-3-methyl-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-1-yl]methyl]-1-bicyclo[2.2.2]octanyl]morpholine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2023-06-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Discovery of the TLR7/8 Antagonist MHV370 for Treatment of Systemic Autoimmune Diseases.
Acs Med.Chem.Lett., 14, 2023
|
|
8DNY
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by decatransin | | Descriptor: | Decatransin peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNZ
 
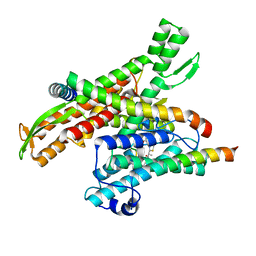 | | Cryo-EM structure of the human Sec61 complex inhibited by apratoxin F | | Descriptor: | Apratoxin F peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DO3
 
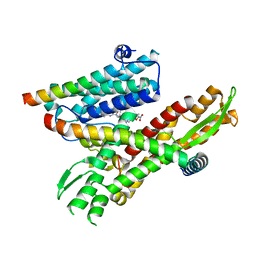 | | Cryo-EM structure of the human Sec61 complex inhibited by eeyarestatin I | | Descriptor: | N'-(4-chlorophenyl)-N-[(4R)-3-(4-chlorophenyl)-5,5-dimethyl-1-(2-{(2E)-2-[(2E)-3-(5-nitrofuran-2-yl)prop-2-en-1-ylidene]hydrazinyl}-2-oxoethyl)-2-oxoimidazolidin-4-yl]-N-hydroxyurea, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNX
 
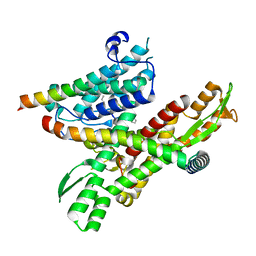 | | Cryo-EM structure of the human Sec61 complex inhibited by cotransin | | Descriptor: | Cotransin analogue peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DNV
 
 | |
8DNW
 
 | |
8DO0
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by mycolactone | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DO1
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
8DO2
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by cyclotriazadisulfonamide (CADA) | | Descriptor: | 9-benzyl-1,5-bis(4-methylbenzene-1-sulfonyl)-3-methylidene-1,5,9-triazacyclododecane, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
4IXD
 
 | | X-ray structure of lfa-1 i-domain in complex with ibe-667 at 1.8a resolution | | Descriptor: | 4-(3-{4-[(3-aminopropyl)carbamoyl]phenyl}-1H-indazol-1-yl)-N-methylbenzamide, Integrin alpha-L, MAGNESIUM ION | | Authors: | Kallen, J. | | Deposit date: | 2013-01-25 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and x-ray structure based investigation of an ICAM-1 binding enhancing small molecule activator of LFA-1
To be Published, 2013
|
|
5XBK
 
 | | Crystal structure of human Importin4 | | Descriptor: | Importin-4, histone H3 | | Authors: | Song, J.J, Yoon, J. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.223 Å) | | Cite: | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
5XAH
 
 | | Crystal structure of human Importin4 | | Descriptor: | Importin-4 | | Authors: | Song, J.J, Yoon, J. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-14 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
6AHC
 
 | |
6JM9
 
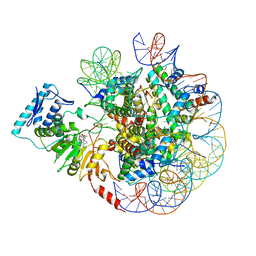 | | cryo-EM structure of DOT1L bound to unmodified nucleosome | | Descriptor: | DNA strand I, DNA strand J, Histone H2A, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
6JMA
 
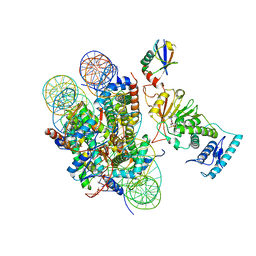 | | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | | Descriptor: | DNA I&J, Histone H2A, Histone H2B 1.1, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
