4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
3MOD
 
 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced IgG) with 11 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3MNZ
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide bearing Ala substitutions in a helical conformation | | Descriptor: | ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, SODIUM ION, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3MNW
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide in a helical conformation | | Descriptor: | 1,2-ETHANEDIOL, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3MOB
 
 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced Fab) with 11 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3MNV
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MO1
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment | | Descriptor: | ACETATE ION, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3MOA
 
 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced Fab) with 17 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
5EA0
 
 | | Structure of the antibody 7968 with human complement factor H-derived peptide | | Descriptor: | Complement factor H-related protein 2, Heavy chain of antibody 7968 Fab fragment, Light chain of antibody 7968 Fab fragment | | Authors: | Bushey, R.T, Moody, M.A, Nicely, N.I, Alam, S.M, Haynes, B.F, Winkler, M.T, Gottlin, E.B, Campa, M.J, Liao, H.-X, Patz Jr, E.F. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Therapeutic Antibody for Cancer, Derived from Single Human B Cells.
Cell Rep, 15, 2016
|
|
4RIS
 
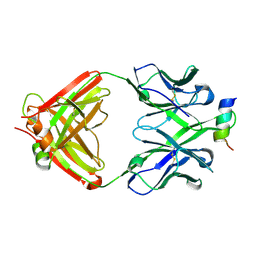 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
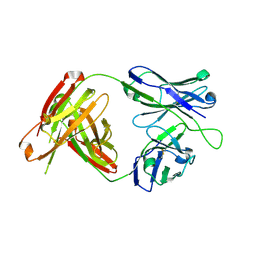 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | Descriptor: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | Authors: | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | Deposit date: | 2014-10-07 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
7LD1
 
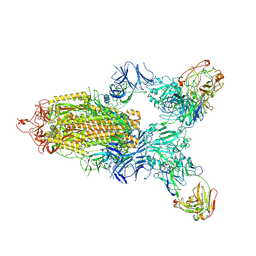 | |
7LCN
 
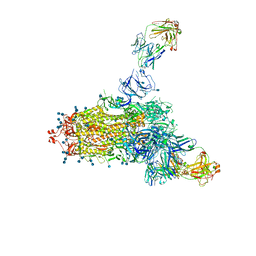 | |
7LAB
 
 | |
7LAA
 
 | |
6MFP
 
 | | Crystal Structure of the RV305 C1-C2 specific ADCC potent antibody DH677.3 Fab in complex with HIV-1 clade A/E gp120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Young, B, Pazgier, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency.
J.Virol., 94, 2020
|
|
5TE4
 
 | | Crystal Structure of Broadly Neutralizing VRC01-class Antibody N6 in Complex with HIV-1 Clade G Strain X2088 gp120 Core | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2016-09-20 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth.
Immunity, 45, 2016
|
|
5T4Z
 
 | |
5KG9
 
 | |
6UDA
 
 | | Cryo-EM structure of CH235UCA bound to Man5-enriched CH505.N279K.G458Y.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH235 UCA heavy chain Fab, CH235 UCA light chain Fab, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancester of CH235 lineage CD4bs broadly neutralizing antibodies.
Plos Pathog., 15, 2019
|
|
6UM5
 
 | | Cryo-EM structure of HIV-1 neutralizing antibody DH270 UCA3 in complex with CH848 10.17DT Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Henderson, R.C, Saunders, K.O, Haynes, B.F. | | Deposit date: | 2019-10-09 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Targeted selection of HIV-specific antibody mutations by engineering B cell maturation.
Science, 366, 2019
|
|
8DY6
 
 | | Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-08-03 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
8DTO
 
 | | Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-07-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
7UPX
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment (local refinement of the RBD and Fab variable domains) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, SP1-77 Fab heavy chain, SP1-77 Fab light chain, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UPY
 
 | | An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
