3I85
 
 | |
3IA6
 
 | |
6XTZ
 
 | |
6YM0
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | Descriptor: | Spike glycoprotein, heavy chain, light chain | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
7JKB
 
 | | 2xVH Fab | | Descriptor: | Anti-Her2, Anti-lysozyme | | Authors: | Lord, D.M, Zhou, Y.F. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Bringing the Heavy Chain to Light: Creating a Symmetric, Bivalent IgG-Like Bispecific.
Antibodies, 9, 2020
|
|
3L7E
 
 | | Crystal structure of ANTI-IL-13 antibody C836 | | Descriptor: | ACETATE ION, C836 HEAVY CHAIN, C836 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen recognition by antibody C836 through adjustment of VL/VH packing
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4KQ4
 
 | | Crystal structure of Anti-IL-17A antibody CNTO7357 | | Descriptor: | CNTO7357 heavy chain, CNTO7357 light chain, NICKEL (II) ION | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-05-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
8BOE
 
 | |
4KQ3
 
 | | Crystal structure of Anti-IL-17A antibody CNTO3186 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CNTO3186 heavy chain, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-05-14 | | Release date: | 2014-03-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural evidence for a constrained conformation of short CDR-L3 in antibodies.
Proteins, 82, 2014
|
|
4KMT
 
 | | Crystal structure of human germline antibody 5-51/O12 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-05-08 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4MAU
 
 | | Crystal structure of anti-ST2L antibody C2244 | | Descriptor: | C2244 heavy chain, C2244 light chain, FORMIC ACID, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4M6O
 
 | | Crystal structure of anti-NGF antibody CNTO7309 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CNTO7309 heavy chain, CNTO7309 light chain, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4M7K
 
 | | Crystal structure of anti-tissue factor antibody 10H10 | | Descriptor: | 10H10 heavy chain, 10H10 light chain, ACETATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4PS4
 
 | | Crystal structure of the complex between IL-13 and M1295 FAB | | Descriptor: | Interleukin-13, M1295 HEAVY CHAIN, M1295 LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2014-03-06 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Framework Adaptation of a Mouse Anti-Human Il-13 Antibody.
J.Mol.Biol., 398, 2010
|
|
6VCA
 
 | | TB38 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
6VC9
 
 | | TB19 complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-nucleotidase, ecto (CD73), ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
7T86
 
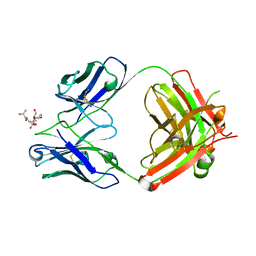 | | Crystal Structure of Fab CR5133 / Phospho-SD Peptide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab CR5133 Heavy Chain, Antibody Fab CR5133 Light Chain, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
7T82
 
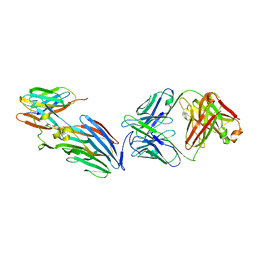 | | Crystal Structure of LEUKOCIDIN E/CENTYRIN S26/FAB B438 | | Descriptor: | Antibody Fab Heavy Chain, Antibody Fab Light Chain, Centyrin S26, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
7T87
 
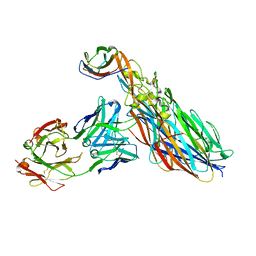 | | CRYSTAL STRUCTURE OF LEUKOCIDIN AB/CENTYRIN S17/FAB 214F COMPLEX | | Descriptor: | Antibody Fab B214 Heavy Chain, Antibody Fab B214 Light Chain, Centyrin S17, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
4DN4
 
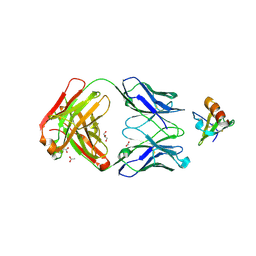 | | Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a | | Descriptor: | ACETATE ION, C-C motif chemokine 2, CNTO888 HEAVY CHAIN, ... | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
1FGV
 
 | |
7JGU
 
 | | Structure of FN3tt mut | | Descriptor: | Fibronectin type-III domain-containing protein, PENTAETHYLENE GLYCOL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
2FGW
 
 | |
5KF4
 
 | |
4DN3
 
 | | Crystal structure of anti-mcp-1 antibody cnto888 | | Descriptor: | CNTO888 HEAVY CHAIN, CNTO888 LIGHT CHAIN | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
