4MSV
 
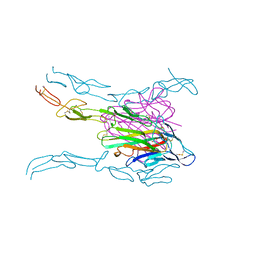 | | Crystal structure of FASL and DcR3 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 6, ... | | Authors: | Liu, W, Ramagopal, U.A, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
7MSJ
 
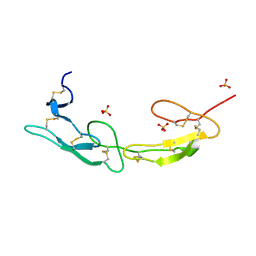 | | The crystal structure of mouse HVEM | | Descriptor: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MSG
 
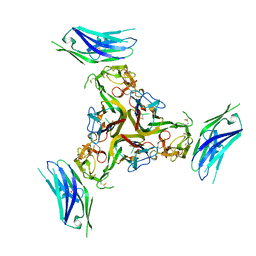 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
3K51
 
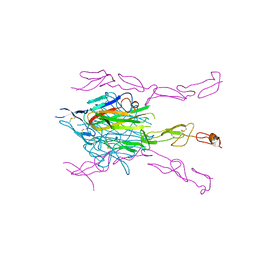 | | Crystal Structure of DcR3-TL1A complex | | Descriptor: | Decoy receptor 3, Tumor necrosis factor ligand superfamily member 15, secreted form | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2009-10-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
4RP3
 
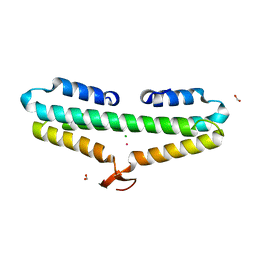 | | Crystal Structure of the L27 Domain of Discs Large 1 (target ID NYSGRC-010766) from Drosophila melanogaster bound to a potassium ion (space group P212121) | | Descriptor: | CHLORIDE ION, Disks large 1 tumor suppressor protein, FORMIC ACID, ... | | Authors: | Ghosh, A, Ramagopal, U, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the L27 Domain of Disc Large Homologue 1 Protein Illustrate a Self-Assembly Module.
Biochemistry, 57, 2018
|
|
2FH7
 
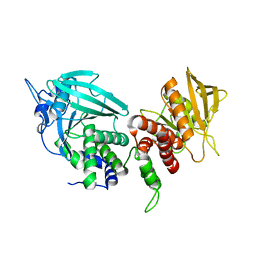 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4RSU
 
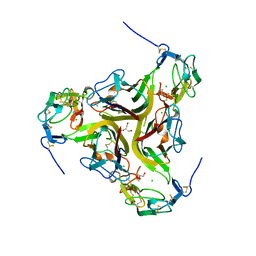 | | Crystal structure of the light and hvem complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, W, Ramagoal, U.A, Himmel, D, Bonanno, J.B, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
1C74
 
 | | Structure of the double mutant (K53,56M) of phospholipase A2 | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Tsai, M.D, Jain, M.K, Ramakumar, S. | | Deposit date: | 2000-01-22 | | Release date: | 2000-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the anionic interface preference and k*cat activation of pancreatic phospholipase A2.
Biochemistry, 39, 2000
|
|
3BP5
 
 | | Crystal structure of the mouse PD-1 and PD-L2 complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Yan, Q, Lazar-Molnar, E, Cao, E, Ramagopal, U.A, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-18 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6NG3
 
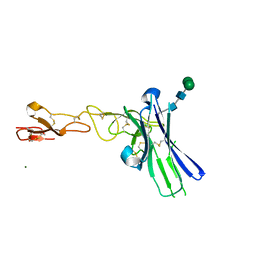 | | Crystal structure of human CD160 and HVEM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | Authors: | Liu, W, Bonanno, J, Almo, S.C. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
6NG9
 
 | |
6NGG
 
 | |
4GOS
 
 | | Crystal structure of human B7-H4 IgV-like domain | | Descriptor: | V-set domain-containing T-cell activation inhibitor 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Vigdorovich, V, Ramagopal, U, Bhosle, R, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and cancer immunotherapy of the B7 family member B7x.
Cell Rep, 9, 2014
|
|
4I0K
 
 | | Crystal structure of murine B7-H3 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD276 antigen, SULFATE ION, ... | | Authors: | Vigdorovich, V, Ramagopal, U, Bonanno, J.B, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structure and T cell inhibition properties of B7 family member, B7-H3.
Structure, 21, 2013
|
|
3BMB
 
 | |
4RP4
 
 | |
4RP5
 
 | |
1RXD
 
 | | Crystal structure of human protein tyrosine phosphatase 4A1 | | Descriptor: | protein tyrosine phosphatase type IVA, member 1; Protein tyrosine phosphatase IVA1 | | Authors: | Sun, J.P, Fedorov, A.A, Almo, S.C, Zhang, Z.Y, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-12-18 | | Release date: | 2004-12-28 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2Q5E
 
 | | Crystal structure of human carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2 | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2Q7O
 
 | | Structure of human purine nucleoside phosphorylase in complex with L-Immucillin-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2007-06-07 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | L-Enantiomers of transition state analogue inhibitors bound to human purine nucleoside phosphorylase.
J.Am.Chem.Soc., 130, 2008
|
|
2HHL
 
 | | Crystal structure of the human small CTD phosphatase 3 isoform 1 | | Descriptor: | 12-TUNGSTOPHOSPHATE, CTD small phosphatase-like protein | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
4ETY
 
 | |
2R0B
 
 | | Crystal structure of human tyrosine phosphatase-like serine/threonine/tyrosine-interacting protein | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine/tyrosine-interacting protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-18 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2QJC
 
 | | Crystal structure of a putative diadenosine tetraphosphatase | | Descriptor: | Diadenosine tetraphosphatase, putative, MANGANESE (II) ION, ... | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-24 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2ZR1
 
 | | Agglutinin from Abrus Precatorius | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Agglutinin-1 chain A, Agglutinin-1 chain B | | Authors: | Cheng, J, Lu, T.H, Liu, C.L, Lin, J.Y. | | Deposit date: | 2008-08-22 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A biophysical elucidation for less toxicity of Agglutinin than Abrin-a from the Seeds of Abrus Precatorius in consequence of crystal structure
J.Biomed.Sci., 17, 2010
|
|
