1HZH
 
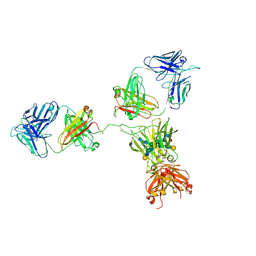 | | CRYSTAL STRUCTURE OF THE INTACT HUMAN IGG B12 WITH BROAD AND POTENT ACTIVITY AGAINST PRIMARY HIV-1 ISOLATES: A TEMPLATE FOR HIV VACCINE DESIGN | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN,Uncharacterized protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Burton, D.R, Wilson, I.A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-08-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design.
Science, 293, 2001
|
|
3M3U
 
 | |
4LOR
 
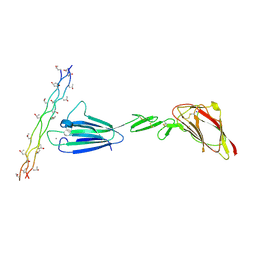 | | C1s CUB1-EGF-CUB2 in complex with a collagen-like peptide from C1q | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1s subcomponent heavy chain, ... | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LMF
 
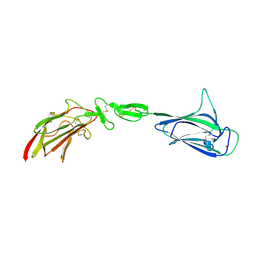 | | C1s CUB1-EGF-CUB2 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain, SODIUM ION | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.921 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LOS
 
 | | C1s CUB2-CCP1 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E, Gingras, A.R. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4LOT
 
 | | C1s CUB2-CCP1-CCP2 | | Descriptor: | Complement C1s subcomponent heavy chain | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-07 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3GW5
 
 | | Crystal structure of human renin complexed with a novel inhibitor | | Descriptor: | (3R)-3-[(1S)-1-(3-chlorophenyl)-1-hydroxy-5-methoxypentyl]-N-{(1S)-2-cyclohexyl-1-[(methylamino)methyl]ethyl}piperidine-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2009-03-31 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and optimization of renin inhibitors: Orally bioavailable alkyl amines.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5CQU
 
 | | Monoclinic Complex Structure of Protein Kinase CK2 Catalytic Subunit with a Benzotriazole-Based Inhibitor Generated by click-chemistry | | Descriptor: | 4-[4-[2-[4,5,6,7-tetrakis(bromanyl)benzotriazol-2-yl]ethyl]-1,2,3-triazol-1-yl]butan-1-amine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Schnitzler, A, Swider, R, Maslyk, M, Ramos, A. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthesis, Biological Activity and Structural Study of New Benzotriazole-Based Protein Kinase CK2 Inhibitors
Rsc Adv, 5, 2015
|
|
5CQW
 
 | | Tetragonal Complex Structure of Protein Kinase CK2 Catalytic Subunit with a Benzotriazole-Based Inhibitor Generated by click-chemistry | | Descriptor: | 4-[4-[2-[4,5,6,7-tetrakis(bromanyl)benzotriazol-2-yl]ethyl]-1,2,3-triazol-1-yl]butan-1-amine, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Schnitzler, A, Swider, R, Maslyk, M, Ramos, A. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis, Biological Activity and Structural Study of New Benzotriazole-Based Protein Kinase CK2 Inhibitors
Rsc Adv, 5, 2015
|
|
5NPS
 
 | | The human O-GlcNAc transferase in complex with a bisubstrate inhibitor | | Descriptor: | 5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] propyl hydrogen phosphate | | Authors: | Rafie, K, van Aalten, D. | | Deposit date: | 2017-04-18 | | Release date: | 2018-05-16 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Thio-Linked UDP-Peptide Conjugates as O-GlcNAc Transferase Inhibitors.
Bioconjug. Chem., 29, 2018
|
|
5NPR
 
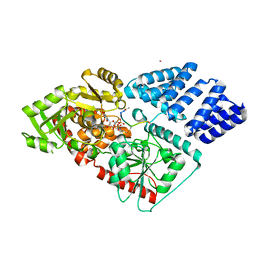 | | The human O-GlcNAc transferase in complex with a thiol-linked bisubstrate inhibitor | | Descriptor: | POTASSIUM ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] propyl hydrogen phosphate, ... | | Authors: | Rafie, K, van Aalten, D.M.F. | | Deposit date: | 2017-04-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thio-Linked UDP-Peptide Conjugates as O-GlcNAc Transferase Inhibitors.
Bioconjug. Chem., 29, 2018
|
|
1UNA
 
 | |
6IAB
 
 | |
1JJ0
 
 | |
6IAC
 
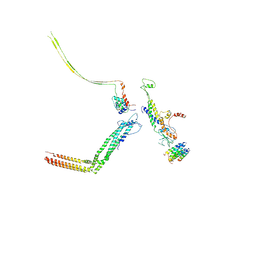 | | Portal and tail of native bacteriophage P68 | | Descriptor: | Lower collar protein, Minor structural protein, Portal protein, ... | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-26 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6IAW
 
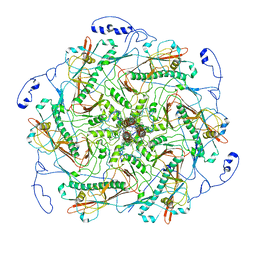 | | Structure of head fiber and inner core protein gp22 of native bacteriophage P68 | | Descriptor: | Arstotzka protein, Head fiber, Major head protein, ... | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
6IAT
 
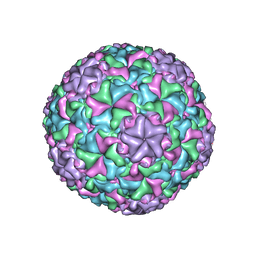 | | Icosahedrally averaged capsid of bacteriophage P68 | | Descriptor: | Arstotzka protein, Major head protein | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
1JIT
 
 | |
6GMA
 
 | |
1JJ3
 
 | |
1JIS
 
 | |
1JIY
 
 | |
6IB1
 
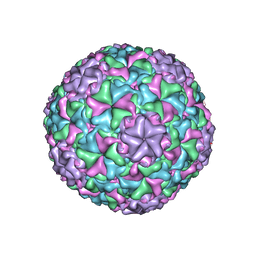 | | Icosahedrally averaged capsid of empty particle of bacteriophage P68 | | Descriptor: | Major head protein, Uncharacterized protein | | Authors: | Hrebik, D, Skubnik, K, Fuzik, T, Plevka, P. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
1JJ1
 
 | |
5M9F
 
 | |
