1G18
 
 | | RECA-ADP-ALF4 COMPLEX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RECA PROTEIN, TETRAFLUOROALUMINATE ION | | Authors: | Datta, S, Prabu, M.M, Vaze, M.B, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation
Nucleic Acids Res., 28, 2000
|
|
1G19
 
 | | STRUCTURE OF RECA PROTEIN | | Descriptor: | PHOSPHATE ION, RECA PROTEIN | | Authors: | Datta, S, Prabu, M.M, Vaze, M.B, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation
Nucleic Acids Res., 28, 2000
|
|
4JMF
 
 | |
1UBF
 
 | | MsREcA-ATPgS complex | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
1UBG
 
 | | MsREcA-dATP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
1UBE
 
 | | MsRecA-ADP Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
1UBC
 
 | | Structure of Reca Protein | | Descriptor: | RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
6JNP
 
 | |
1MO4
 
 | |
1MO5
 
 | |
1MO6
 
 | |
1MO3
 
 | |
1PBY
 
 | | Structure of the Phenylhydrazine Adduct of the Quinohemoprotein Amine Dehydrogenase from Paracoccus denitrificans at 1.7 A Resolution | | Descriptor: | HEME C, TERTIARY-BUTYL ALCOHOL, quinohemoprotein amine dehydrogenase 40 kDa subunit, ... | | Authors: | Datta, S, Ikeda, T, Kano, K, Mathews, F.S. | | Deposit date: | 2003-05-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the phenylhydrazine adduct of the quinohemoprotein amine dehydrogenase from Paracoccus denitrificans at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P4D
 
 | | F factor TraI Relaxase Domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TraI protein | | Authors: | Datta, S, Larkin, C, Schildbach, J.F. | | Deposit date: | 2003-04-22 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into single-stranded DNA binding and cleavage by F factor TraI.
Structure, 11, 2003
|
|
1JJU
 
 | | Structure of a Quinohemoprotein Amine Dehydrogenase with a Unique Redox Cofactor and Highly Unusual Crosslinking | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, QUINOHEMOPROTEIN AMINE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Datta, S, Mori, Y, Takagi, K, Kawaguchi, K, Chen, Z.-W, Kano, K, Ikeda, T, Okajima, T, Kuroda, S, Tanizawa, K, Mathews, F.S. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JIT
 
 | |
1JIS
 
 | |
1JIY
 
 | |
1JJ3
 
 | |
1JJ1
 
 | |
1JJ0
 
 | |
8HUC
 
 | | Crystal structure of PaIch (Pec1) | | Descriptor: | GLYCEROL, MaoC_dehydrat_N domain-containing protein, NITRATE ION | | Authors: | Pramanik, A, Datta, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa Effector Candidate 1(Pec1).
To Be Published
|
|
7EVA
 
 | |
4RUK
 
 | |
5C1T
 
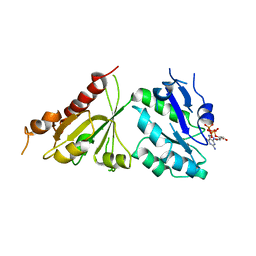 | | Crystal structure of the GTP-bound wild type EhRabX3 from Entamoeba histolytica | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Small GTPase EhRabX3 | | Authors: | Srivastava, V.K, Chandra, M, Datta, S. | | Deposit date: | 2015-06-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal Structure Analysis of Wild Type and Fast Hydrolyzing Mutant of EhRabX3, a Tandem Ras Superfamily GTPase from Entamoeba histolytica.
J.Mol.Biol., 428, 2016
|
|
