2KT8
 
 | | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B | | Descriptor: | Probable surface protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B
To be Published
|
|
2LXU
 
 | | Solution NMR Structure of the eukaryotic RNA recognition motif, RRM1, from the heterogeneous nuclear ribonucleoprotein H from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8614A | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Ramelot, T.A, Yang, Y, Pederson, K, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the eukaryotic RNA recognition motif, RRM1, from the heterogeneous nuclear ribonucleoprotein H from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8614A
To be Published
|
|
2MK2
 
 | | Solution NMR structure of N-terminal domain (SH2 domain) of human Inositol polyphosphate phosphatase-like protein 1 (INPPL1) (fragment 20-117), Northeast Structural Genomics Consortium Target HR9134A | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Yang, Y, Ramelot, T.A, Janjua, H, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-01-23 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of N-terminal domain (SH2 domain) of human Inositol polyphosphate phosphatase-like protein 1 (INPPL1) (fragment 20-117), Northeast Structural Genomics Consortium Target HR9134A.
To be Published
|
|
2MA6
 
 | | Solution NMR Structure of the RING finger domain from the Kip1 ubiquitination-promoting E3 complex protein 1 (KPC1/RNF123) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8700A | | Descriptor: | E3 ubiquitin-protein ligase RNF123, ZINC ION | | Authors: | Ramelot, T.A, Yang, Y, Janjua, H, Kohan, E, Wang, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the RING finger domain from the Kip1 ubiquitination-promoting E3 complex protein 1 (KPC1/RNF123) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8700A
To be Published
|
|
6JIQ
 
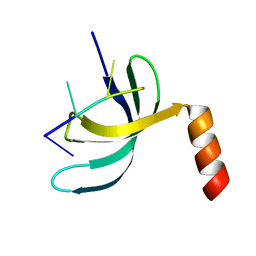 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIP
 
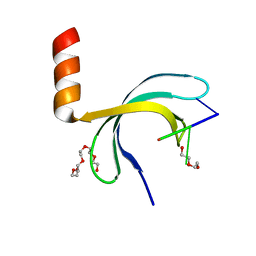 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | Authors: | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5TVW
 
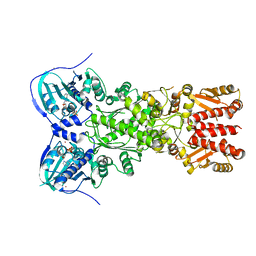 | | Crystal structure of mitochondrial Hsp90 (TRAP1) with ATP in absence of Mg, hemi-hydrolyzed | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Elnatan, D, Betegon, M, Agard, D.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-11-10 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Symmetry broken and rebroken during the ATP hydrolysis cycle of the mitochondrial Hsp90 TRAP1.
Elife, 6, 2017
|
|
1TVG
 
 | | X-ray structure of human PP25 gene product, HSPC034. Northeast Structural Genomics Target HR1958. | | Descriptor: | CALCIUM ION, LOC51668 protein, SAMARIUM (III) ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
8GLT
 
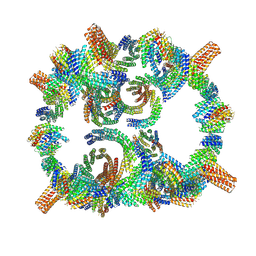 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | Descriptor: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | Authors: | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
2AYJ
 
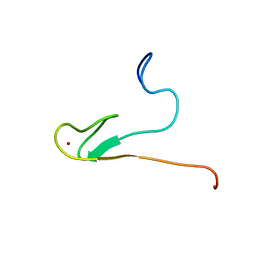 | | Solution structure of 50S ribosomal protein L40e from Sulfolobus solfataricus | | Descriptor: | 50S ribosomal protein L40e, ZINC ION | | Authors: | Wu, B, Yee, A, Lukin, J, Lemak, A, Semesi, A, Ramelot, T, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-07 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein L40E, a unique C4 zinc finger protein encoded by archaeon Sulfolobus solfataricus
Protein Sci., 17, 2008
|
|
6XR1
 
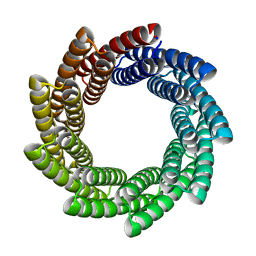 | |
6XR2
 
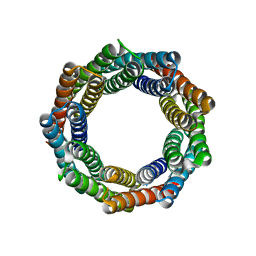 | |
1EJE
 
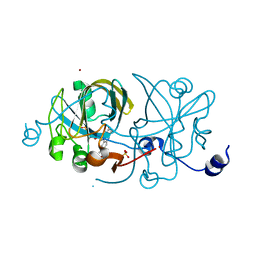 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
1EIJ
 
 | | NMR ENSEMBLE OF METHANOBACTERIUM THERMOAUTOTROPHICUM PROTEIN 1615 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1615 | | Authors: | Christendat, D, Booth, V, Gernstein, M, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
3H9X
 
 | | Crystal Structure of the PSPTO_3016 protein from Pseudomonas syringae, Northeast Structural Genomics Consortium Target PsR293 | | Descriptor: | uncharacterized protein PSPTO_3016 | | Authors: | Seetharaman, J, Lew, S, Forouhar, F, Hamilton, H, Xiao, R, Ciccosanti, C, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Solution NMR and X-ray crystal structures of Pseudomonas syringae Pspto_3016 from protein domain family PF04237 (DUF419) adopt a "double wing" DNA binding motif.
J.Struct.Funct.Genom., 13, 2012
|
|
7MIU
 
 | | Mouse CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIG
 
 | | Human CTPS1 bound to inhibitor T35 | | Descriptor: | 2-{2-[(cyclopropanesulfonyl)amino]-1,3-thiazol-4-yl}-2-methyl-N-{5-[6-(trifluoromethyl)pyrazin-2-yl]pyridin-2-yl}propanamide, CTP synthase 1, GLUTAMINE, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIH
 
 | | Human CTPS2 bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIF
 
 | | Human CTPS1 bound to inhibitor R80 | | Descriptor: | CTP synthase 1, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIV
 
 | | Mouse CTPS2-I250T bound to inhibitor R80 | | Descriptor: | CTP synthase 2, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MII
 
 | | Human CTPS2 bound to inhibitor T35 | | Descriptor: | 2-{2-[(cyclopropanesulfonyl)amino]-1,3-thiazol-4-yl}-2-methyl-N-{5-[6-(trifluoromethyl)pyrazin-2-yl]pyridin-2-yl}propanamide, CTP synthase 2, GLUTAMINE, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MIP
 
 | | Mouse CTPS1 bound to inhibitor R80 | | Descriptor: | CTP synthase 1, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MGZ
 
 | | Human CTPS1 bound to UTP, AMPPNP, and glutamine | | Descriptor: | CTP synthase 1, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Lynch, E.M, Dimattia, M.A, Kollman, J.M. | | Deposit date: | 2021-04-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for isoform-specific inhibition of human CTPS1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5ZKL
 
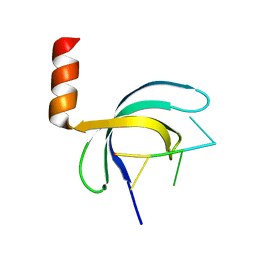 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5ZKM
 
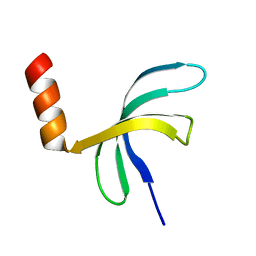 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA TCTTCC | | Descriptor: | DNA (5'-D(P*TP*CP*TP*TP*CP*C)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
