4IFT
 
 | | Crystal structure of double mutant thermostable NPPase from Geobacillus stearothermophilus | | Descriptor: | Thermostable NPPase | | Authors: | Guo, Z, Huang, J, Wang, F, Qiu, R, Wang, Y, Ji, C. | | Deposit date: | 2012-12-15 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Crystal structure of thermostable NPPase from Geobacillus stearothermophilus
To be Published
|
|
5BZ6
 
 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
3QYC
 
 | | Structure of a dimeric anti-HER2 single domain antibody | | Descriptor: | VH domain of IgG molecule | | Authors: | Baral, T.N, Chao, S, Li, S, Tanha, J, Arbabai, M, Wang, S, Zhang, J. | | Deposit date: | 2011-03-03 | | Release date: | 2012-02-08 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Human Single Domain Antibody Dimer Formed through V(H)-V(H) Non-Covalent Interactions.
Plos One, 7, 2012
|
|
3RLR
 
 | | Co-crystal structure of the HSP90 ATP binding domain in complex with 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile | | Descriptor: | 4-(2,4-dichloro-5-methoxyphenyl)-2,6-dimethyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Kung, P.-P, Sinnema, P.-J, Richardson, P, Hickey, M.J, Gajiwala, K.S, Wang, F, Huang, B, McClellan, G, Wang, J, Maegley, K, Bergqvist, S, Mehta, P.P, Kania, R. | | Deposit date: | 2011-04-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design strategies to target crystallographic waters applied to the Hsp90 molecular chaperone.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2HQ4
 
 | | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH1570 | | Authors: | Li, Y, Marshall, M, Chang, J, Zhao, M, Zhang, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii
To be Published
|
|
4LMQ
 
 | | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 | | Descriptor: | Stromal cell-derived factor 1, hu30D8 Fab heavy chain, hu30D8 Fab light chain | | Authors: | Zhong, Z, Wang, J, Li, B, Xiang, H, Ultsch, M, Coons, M, Wong, T, Chiang, N.Y, Clark, S, Clark, R, Quintana, L, Gribling, P, Suto, E, Barck, K, Corpuz, R, Yao, J, Takkar, R, Lee, W.P, Damico-Beyer, L.A, Carano, R.D, Adams, C, Kelley, R.F, Wang, W, Ferrara, N. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12.
Clin.Cancer Res., 19, 2013
|
|
3NQZ
 
 | | Crystal structure of the autoprocessed Vibriolysin MCP-02 with E369A mutation | | Descriptor: | CALCIUM ION, Secreted metalloprotease Mcp02, ZINC ION | | Authors: | Gao, X, Wang, J, Chen, L, Wu, J.-W, Zhang, Y.-Z. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6JPD
 
 | | Mouse receptor-interacting protein kinase 3 (RIP3) amyloid structure by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Hu, H, Zhang, J, Dong, X.Q, Wang, J, Schwieters, C, Wang, H.Y, Lu, J.X. | | Deposit date: | 2019-03-26 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis.
Nat Commun, 12, 2021
|
|
5ZAL
 
 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
7WKH
 
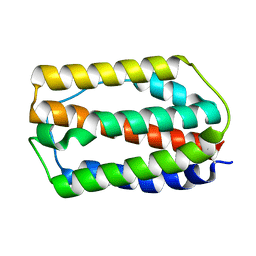 | | the grass carp IFNa | | Descriptor: | Interferon | | Authors: | Zou, J, WANG, J, Wang, Z. | | Deposit date: | 2022-01-09 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Functional Analyses of Type I IFNa Shed Light Into Its Interaction With Multiple Receptors in Fish.
Front Immunol, 13, 2022
|
|
3NQY
 
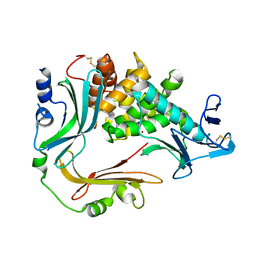 | | Crystal structure of the autoprocessed complex of Vibriolysin MCP-02 with a single point mutation E346A | | Descriptor: | CALCIUM ION, Secreted metalloprotease Mcp02, ZINC ION | | Authors: | Gao, X, Wang, J, Wu, J.-W, Zhang, Y.-Z. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the autoprocessing of zinc metalloproteases in the thermolysin family
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4EAJ
 
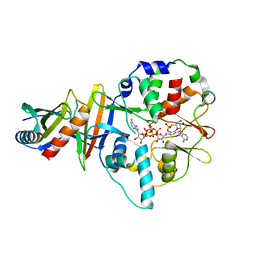 | | Co-crystal of AMPK core with AMP soaked with ATP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
7CLT
 
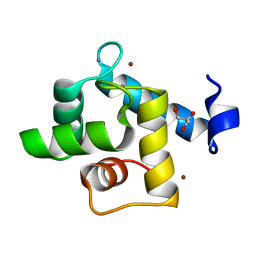 | | Crystal structure of the EFhd1/Swiprosin-2, a mitochondrial actin-binding protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D1, GLYCEROL, ... | | Authors: | Mun, S.A, Park, J, Park, K.R, Lee, Y, Kang, J.Y, Park, T, Jin, M, Yang, J, Jun, C.D, Eom, S.H. | | Deposit date: | 2020-07-22 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07380986 Å) | | Cite: | Structural and Biochemical Characterization of EFhd1/Swiprosin-2, an Actin-Binding Protein in Mitochondria.
Front Cell Dev Biol, 8, 2020
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
6Z71
 
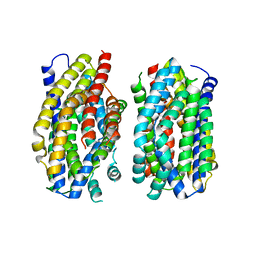 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z70
 
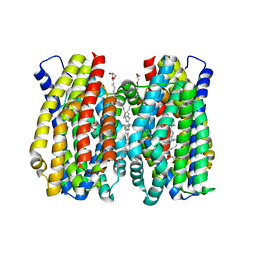 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4EQA
 
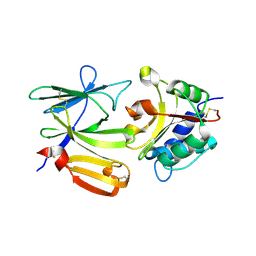 | | Crystal structure of PA1844 in complex with PA1845 from Pseudomonas aeruginosa PAO1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
7LW2
 
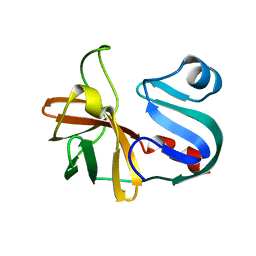 | |
6JUI
 
 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | Descriptor: | Pre-mRNA-splicing factor CEF1 | | Authors: | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | Deposit date: | 2019-04-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
2P5B
 
 | | The complex structure of JMJD2A and trimethylated H3K36 peptide | | Descriptor: | FE (II) ION, Histone H3, JmjC domain-containing histone demethylation protein 3A, ... | | Authors: | Zhang, G, Chen, Z, Zang, J, Hong, X, Shi, Y. | | Deposit date: | 2007-03-14 | | Release date: | 2007-06-12 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
5GN8
 
 | | Structure of a 48-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | CALCIUM ION, Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Zhao, G, Mikami, B. | | Deposit date: | 2016-07-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | "Silent" Amino Acid Residues at Key Subunit Interfaces Regulate the Geometry of Protein Nanocages
ACS Nano, 10, 2016
|
|
4F2L
 
 | | Structure of a regulatory domain of AMPK | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, MAGNESIUM ION | | Authors: | Xin, F.J, Zhang, Y.Y, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2012-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved regulatory elements in AMPK
Nature, 498, 2013
|
|
8J22
 
 | | Cryo-EM structure of FFAR2 complex bound with TUG-1375 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J20
 
 | | Cryo-EM structure of FFAR3 bound with valeric acid and AR420626 | | Descriptor: | (4R)-N-[2,5-bis(chloranyl)phenyl]-4-(furan-2-yl)-2-methyl-5-oxidanylidene-4,6,7,8-tetrahydro-1H-quinoline-3-carboxamide, Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J24
 
 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | Descriptor: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
