3UNG
 
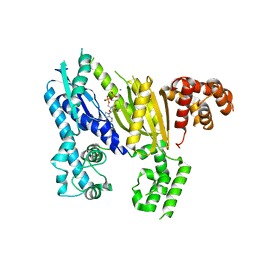 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Cmr2dHD, ... | | Authors: | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
3V5S
 
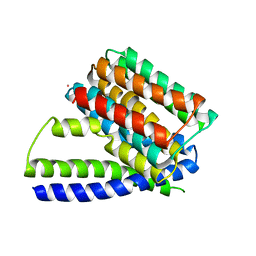 | | Structure of Sodium/Calcium Exchanger from Methanococcus jannaschii | | Descriptor: | CADMIUM ION, Sodium/Calcium Exchanger | | Authors: | Liao, J, Li, H, Zeng, W, Sauer, D.B, Belmares, R, Jiang, Y. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Science, 335, 2012
|
|
4NV3
 
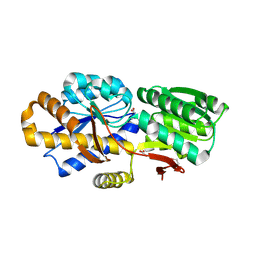 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with valine. | | Descriptor: | ACETATE ION, Amino acid/amide ABC transporter substrate-binding protein, HAAT family, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with valine.
To be Published
|
|
4NSE
 
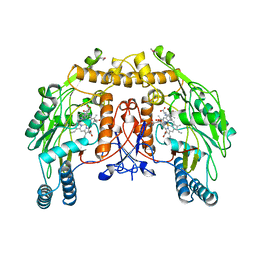 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, L-ARG COMPLEX | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1998-10-07 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Cell(Cambridge,Mass.), 95, 1998
|
|
4APP
 
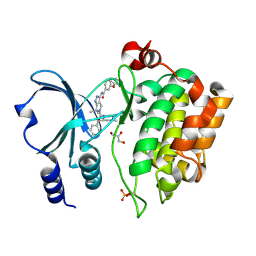 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with (S)-N-(5-(3-benzyl-1-methylpiperazine-4-carbonyl)-6,6-dimethyl-1,4,5, 6-tetrahydropyrrolo(3,4-c)pyrazol-3-yl)-3-phenoxybenzamide | | Descriptor: | GLYCEROL, N-[6,6-dimethyl-5-[(2S)-4-methyl-2-(phenylmethyl)piperazin-1-yl]carbonyl-2,4-dihydropyrrolo[3,4-c]pyrazol-3-yl]-3-phenoxy-benzamide, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.D, Deng, Y.L, Wang, C, Guo, C, McAlpine, I, Zhang, J, Kephart, S, Johnson, M.C, Li, H, Bouzida, D, Yang, A, Dong, L, Marakovits, J, Tikhe, J, Richardson, P, Guo, L.C, Kania, R, Edwards, M.P, Kraynov, E, Christensen, J, Piraino, J, Lee, J, Dagostino, E, Del-Carmen, C, Smeal, T, Murray, B.W. | | Deposit date: | 2012-04-04 | | Release date: | 2012-06-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Pyrroloaminopyrazoles as Novel Pak Inhibitors.
J.Med.Chem., 55, 2012
|
|
4MOZ
 
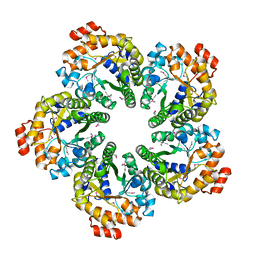 | |
2GJW
 
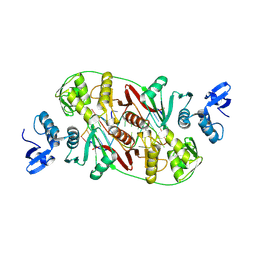 | | RNA Recognition and Cleavage by an Splicing Endonuclease | | Descriptor: | 5'-R(*AP*GP*GP*UP*CP*GP*C)-3', 5'-R(*GP*CP*GP*AP*CP*CP*GP*AP*CP*CP*AP*(DU)P*AP*GP*CP*UP*GP*CP*A)-3', 5'-R(*UP*GP*CP*AP*GP*CP*GP*GP*UP*CP*AP*(A23))-3', ... | | Authors: | Xue, S, Calvin, K, Li, H. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | RNA Recognition and Cleavage by an Splicing Endonuclease
Science, 312, 2006
|
|
3UR3
 
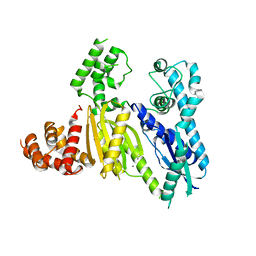 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | Descriptor: | CALCIUM ION, Cmr2dHD, ZINC ION | | Authors: | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
4NW9
 
 | |
3UPS
 
 | |
3V5U
 
 | | Structure of Sodium/Calcium Exchanger from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ACETATE ION, CALCIUM ION, ... | | Authors: | Jiang, Y, Liao, J, Li, H, Zeng, W, Sauer, D, Belmares, R. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger.
Science, 335, 2012
|
|
4LS0
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH01B0033 | | Descriptor: | 2-{(E)-[2-(4-phenyl-1,3-thiazol-2-yl)hydrazinylidene]methyl}benzaldehyde, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DHO1B0033
To be Published
|
|
4NTP
 
 | |
2I6X
 
 | | The structure of a predicted HAD-like family hydrolase from Porphyromonas gingivalis. | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Cuff, M.E, Mussar, K.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-29 | | Release date: | 2006-10-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of a predicted HAD-like family hydrolase from Porphyromonas gingivalis. (CASP Target)
To be Published
|
|
6IMA
 
 | | Crystal Structure of ALKBH1 without alpha-1 (N37-C369) | | Descriptor: | CITRIC ACID, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
4NQR
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with alanine | | Descriptor: | ALANINE, Amino acid/amide ABC transporter substrate-binding protein, HAAT family, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-11-25 | | Release date: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with alanine.
To be Published
|
|
2GS8
 
 | | Structure of mevalonate pyrophosphate decarboxylase from Streptococcus pyogenes | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, mevalonate pyrophosphate decarboxylase | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of mevalonate pyrophosphate decarboxylase from Streptococcus pyogenes
To be Published
|
|
4NTR
 
 | |
4NW8
 
 | |
4O5A
 
 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
2HKX
 
 | | Structure of CooA mutant (N127L/S128L) from Carboxydothermus hydrogenoformans | | Descriptor: | CARBON MONOXIDE, Carbon monoxide oxidation system transcription regulator CooA-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lanz, N.D, Borjigin, M, Li, H, Kerby, R.L, Poulos, T.L, Roberts, G.P. | | Deposit date: | 2006-07-05 | | Release date: | 2007-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based hypothesis on the activation of the CO-sensing transcription factor CooA.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2I0M
 
 | | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae | | Descriptor: | Phosphate transport system protein phoU, ZINC ION | | Authors: | Zhang, R, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-10 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the phosphate transport system regulatory protein PhoU from Streptococcus pneumoniae
To be Published, 2006
|
|
2HVC
 
 | | The Crystal Structure of Ligand-binding Domain (LBD) of human Androgen Receptor in Complex with a selective modulator LGD2226 | | Descriptor: | 6-[BIS(2,2,2-TRIFLUOROETHYL)AMINO]-4-(TRIFLUOROMETHYL)QUINOLIN-2(1H)-ONE, Androgen receptor | | Authors: | Wang, F, Liu, X.-Q, Li, H, Liang, K.-N, Miner, J.N, Hong, M, Kallel, E.A, van Oeveren, A, Zhi, L, Jiang, T. | | Deposit date: | 2006-07-28 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ligand-binding domain (LBD) of human androgen receptor in complex with a selective modulator LGD2226
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2G2C
 
 | |
4AZN
 
 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag-mediated crystal packing | | Descriptor: | FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
