1LRY
 
 | | Crystal Structure of P. aeruginosa Peptide Deformylase Complexed with Antibiotic Actinonin | | Descriptor: | ACTINONIN, PEPTIDE deformylase, ZINC ION | | Authors: | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | Deposit date: | 2002-05-16 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
1V6N
 
 | | Peanut lectin with 9mer peptide (PVIWSSATG) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION | | Authors: | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-12-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2Y6C
 
 | | The Discovery of MMP7 inhibitors Exploiting a Novel Selectivity Trigger | | Descriptor: | CALCIUM ION, MATRILYSIN, N-{[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]SULFONYL}-L-TRYPTOPHAN, ... | | Authors: | Edman, K, Furber, M, Hemsley, P, Johansson, C, Pairaudeau, G, Petersen, J, Stocks, M, Tervo, A, Ward, A, Wells, E, Wissler, L. | | Deposit date: | 2011-01-20 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Discovery of Mmp7 Inhibitors Exploiting a Novel Selectivity Trigger.
Chemmedchem, 6, 2011
|
|
1UXC
 
 | | FRUCTOSE REPRESSOR DNA-BINDING DOMAIN, NMR, MINIMIZED STRUCTURE | | Descriptor: | FRUCTOSE REPRESSOR | | Authors: | Penin, F, Geourjon, C, Montserret, R, Bockmann, A, Lesage, A, Yang, Y, Bonod-Bidaud, C, Cortay, J.C, Negre, D, Cozzone, A.J, Deleage, G. | | Deposit date: | 1996-12-26 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR.
J.Mol.Biol., 270, 1997
|
|
2YFO
 
 | | GALACTOSIDASE DOMAIN OF ALPHA-GALACTOSIDASE-SUCROSE KINASE, AGASK, in complex with galactose | | Descriptor: | ALPHA-GALACTOSIDASE-SUCROSE KINASE AGASK, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sulzenbacher, G, Bruel, L, Tison-Cervera, M, Pujol, A, Nicoletti, C, Perrier, J, Galinier, A, Ropartz, D, Fons, M, Pompeo, F, Giardina, T. | | Deposit date: | 2011-04-07 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Agask, a Bifunctional Enzyme from the Human Microbiome Coupling Galactosidase and Kinase Activities
J.Biol.Chem., 286, 2011
|
|
2YLS
 
 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: REDUCED ENZYME BOUND TO NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLACETONE MONOOXYGENASE | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
1NCT
 
 | | TITIN MODULE M5, N-TERMINALLY EXTENDED, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
1URI
 
 | | AZURIN MUTANT WITH MET 121 REPLACED BY GLN | | Descriptor: | AZURIN, COPPER (II) ION, SULFATE ION | | Authors: | Romero, A, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1996-11-14 | | Release date: | 1997-04-01 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray analysis and spectroscopic characterization of M121Q azurin. A copper site model for stellacyanin.
J.Mol.Biol., 229, 1993
|
|
3PRW
 
 | | Crystal structure of the lipoprotein BamB | | Descriptor: | Lipoprotein yfgL | | Authors: | Heuck, A, Clausen, T. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Augmenting beta-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins.
J.Mol.Biol., 406, 2011
|
|
2QL3
 
 | | Crystal structure of the C-terminal domain of a probable LysR family transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | PHOSPHATE ION, Probable transcriptional regulator, LysR family protein | | Authors: | Tan, K, Skarina, T, Kagen, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-12 | | Release date: | 2007-07-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of the C-terminal domain of a probable LysR family transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3PSY
 
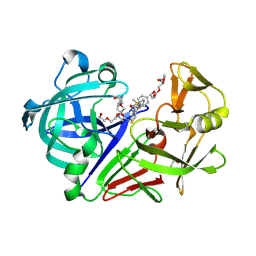 | | Endothiapepsin in complex with an inhibitor based on the Gewald reaction | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-12-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1EOS
 
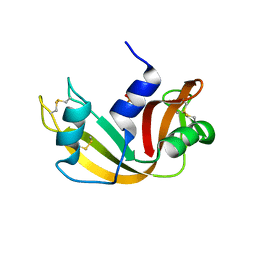 | | CRYSTAL STRUCTURE OF RIBONUCLEASE A COMPLEXED WITH URIDYLYL(2',5')GUANOSINE (PRODUCTIVE BINDING) | | Descriptor: | RIBONUCLEASE PANCREATIC, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 2000-03-24 | | Release date: | 2000-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Productive and nonproductive binding to ribonuclease A: X-ray structure of two complexes with uridylyl(2',5')guanosine.
Protein Sci., 9, 2000
|
|
2YHD
 
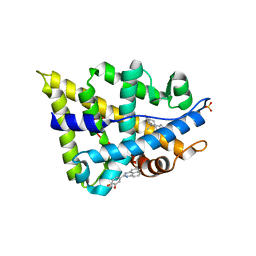 | | Human androgen receptor in complex with AF2 small molecule inhibitor | | Descriptor: | 4-(2,3-dihydro-1H-perimidin-2-yl)benzene-1,2-diol, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | AxerioCilies, P, Lack, N.A, ShashiNayana, M.R, Chan, K.H, Yeung, A, LeBlanc, E, Guns, E, Rennie, P, Cherkasov, A. | | Deposit date: | 2011-04-28 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Androgen Receptor Activation Function-2 (Af2) Site Identified Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
4TY7
 
 | |
1NJM
 
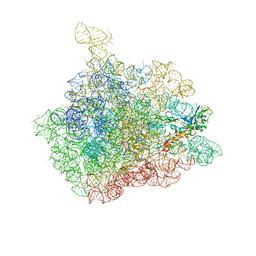 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) and the antibiotic sparsomycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
5Z5I
 
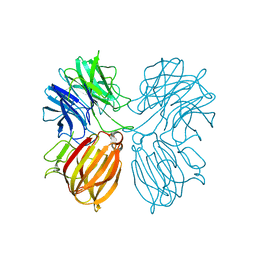 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose and D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose, ... | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
2YM2
 
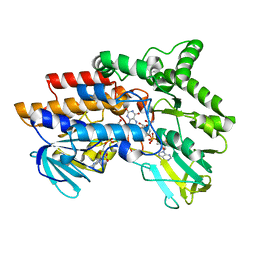 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: Arg337Lys MUTANT REDUCED STATE WITH NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLACETONE MONOOXYGENASE | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
1N9G
 
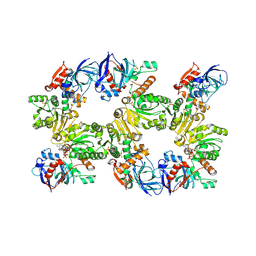 | | Mitochondrial 2-enoyl thioester reductase Etr1p/Etr2p heterodimer from Candida tropicalis | | Descriptor: | 2,4-dienoyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Torkko, J.M, Koivuranta, K.T, Kastaniotis, A.J, Airenne, T.T, Glumoff, T, Ilves, M, Hartig, A, Gurvitz, A, Hiltunen, J.K. | | Deposit date: | 2002-11-25 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Candida tropicalis expresses two mitochondrial 2-enoyl thioester reductases that are able to form both homodimers and heterodimers.
J.Biol.Chem., 278, 2003
|
|
1NL3
 
 | | CRYSTAL STRUCTURE OF THE SECA PROTEIN TRANSLOCATION ATPASE FROM MYCOBACTERIUM TUBERCULOSIS in APO FORM | | Descriptor: | PREPROTEIN TRANSLOCASE SECA 1 SUBUNIT | | Authors: | Sharma, V, Arockiasamy, A, Ronning, D.R, Savva, C.G, Holzenburg, A, Braunstein, M, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of M. tuberculosis SecA, A Preprotein Translocating ATPase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1URG
 
 | | X-ray structures from the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
2LEZ
 
 | | Solution NMR structure of N-terminal domain of Salmonella effector protein PipB2. Northeast structural genomics consortium (NESG) target stt318a | | Descriptor: | Secreted effector protein pipB2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Daniels, C, Savchenko, A, Arrowsmith, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of N-terminal domain of Salmonella effector protein PipB2
To be Published
|
|
4RWA
 
 | | Synchrotron structure of the human delta opioid receptor in complex with a bifunctional peptide (PSI community target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Soluble cytochrome b562,Delta-type opioid receptor, bifunctional peptide | | Authors: | Fenalti, G, Zatsepin, N.A, Betti, C, Giguere, P, Han, G.W, Ishchenko, A, Liu, W, Guillemyn, K, Zhang, H, James, D, Wang, D, Weierstall, U, Spence, J.C.H, Boutet, S, Messerschmidt, M, Williams, G.J, Gati, C, Yefanov, O.M, White, T.A, Oberthuer, D, Metz, M, Yoon, C.H, Barty, A, Chapman, H.N, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Fromme, P, Tourwe, D, Schiller, P.W, Roth, B.L, Ballet, S, Katritch, V, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2014-12-01 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for bifunctional peptide recognition at human delta-opioid receptor.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4S39
 
 | | IspG in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, GLYCEROL, ... | | Authors: | Quitterer, F, Frank, A, Wang, K, Guodong, R, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
2LS8
 
 | | Solution structure of human C-type lectin domain family 4 member D | | Descriptor: | C-type lectin domain family 4 member D | | Authors: | Harris, R, Gaudette, J, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human C-type lectin domain family 4 member D
To be Published
|
|
2LDA
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP2 (Ac-HKXLHQXLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP2 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
