3ZWN
 
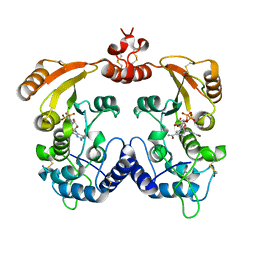 | | Crystal structure of Aplysia cyclase complexed with substrate NGD and product cGDPR | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, CYCLIC GUANOSINE DIPHOSPHATE-RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
2OD3
 
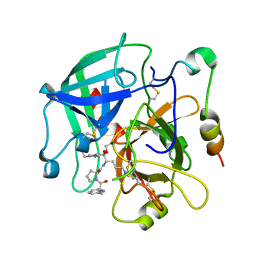 | | Human thrombin chimera with human residues 184a, 186, 186a, 186b, 186c and 222 replaced by murine thrombin equivalents. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Thrombin heavy chain, ... | | Authors: | Marino, F, Chen, Z, Ergenekan, C.E, Bush, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of na+ activation mimicry in murine thrombin.
J.Biol.Chem., 282, 2007
|
|
4HYM
 
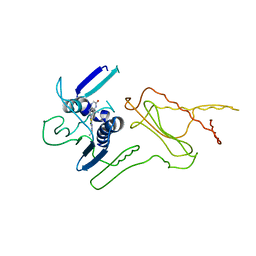 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)pyrido[2,3-b]pyrazin-2(1H)-one, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6K3H
 
 | |
4KYT
 
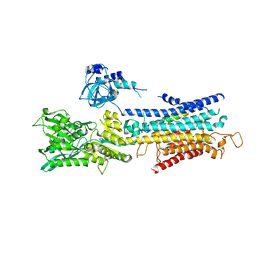 | | The structure of superinhibitory phospholamban bound to the calcium pump SERCA1a | | Descriptor: | Cardiac phospholamban, POTASSIUM ION, SERCA1a, ... | | Authors: | Hurley, T.D, Akin, B.L, Jones, L.R. | | Deposit date: | 2013-05-29 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum.
J.Biol.Chem., 288, 2013
|
|
3ZWV
 
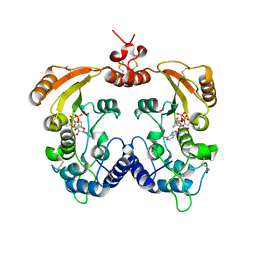 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of intermediates along the cyclization pathway of Aplysia ADP-ribosyl cyclase.
J. Mol. Biol., 415, 2012
|
|
3ZWP
 
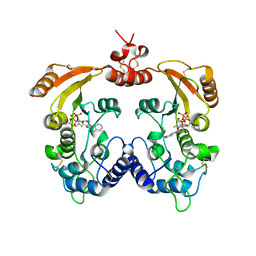 | | Crystal structure of ADP ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.1 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, GLYCEROL, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
7WE6
 
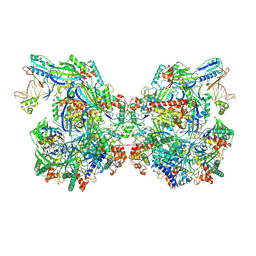 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
4LCW
 
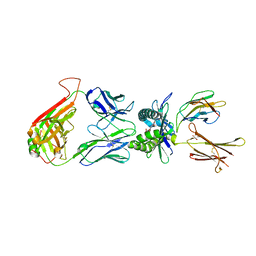 | | The structure of human MAIT TCR in complex with MR1-K43A-RL-6-Me-7OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells.
J.Exp.Med., 210, 2013
|
|
4NA5
 
 | |
3OY5
 
 | |
3OX7
 
 | | The crystal structure of uPA complex with peptide inhibitor MH027 at pH4.6 | | Descriptor: | MH027, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Andreasen, P.A, Huang, M.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The binding mechanism of a peptidic cyclic serine protease inhibitor
J.Mol.Biol., 412, 2011
|
|
3OOI
 
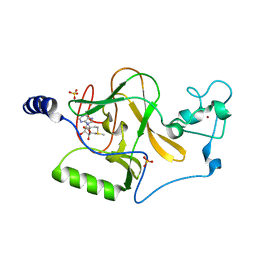 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Qiao, Q, Wang, M, Xu, R.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
5K7X
 
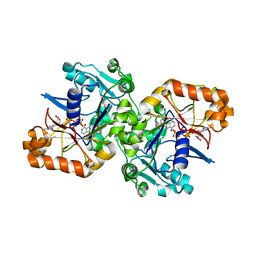 | |
3ROP
 
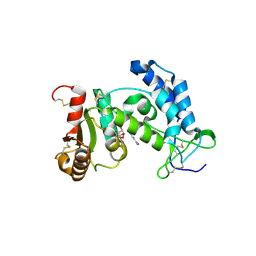 | |
3ROQ
 
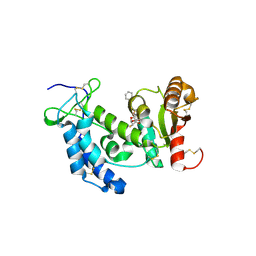 | | Crystal structure of human CD38 in complex with compound CZ-46 | | Descriptor: | (2R,3R,4S,5S)-4-fluoro-3,5-dihydroxytetrahydrofuran-2-yl 2-phenylethyl hydrogen (S)-phosphate, ADP-ribosyl cyclase 1 | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-04-26 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis-based inhibitors of the calcium signaling function of CD38.
Biochemistry, 51, 2012
|
|
3ROM
 
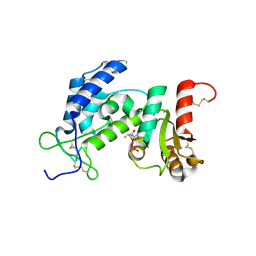 | |
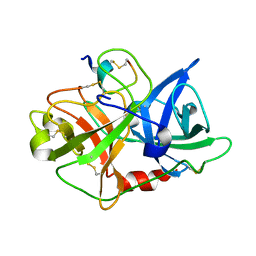
3OY6
 
 | |
7WX6
 
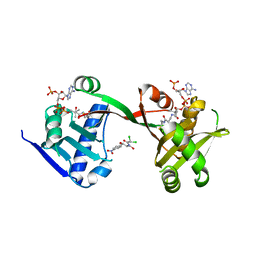 | | A Legionella acetyltransferase VipF | | Descriptor: | CHLORAMPHENICOL, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Chen, Z, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3ROK
 
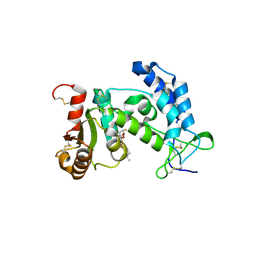 | |
7FJ1
 
 | | Cryo-EM structure of pseudorabies virus C-capsid | | Descriptor: | Capsid vertex component 1, DNA packaging tegument protein UL25, Major capsid protein, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Sun, H. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
7FJ3
 
 | | Cryo-EM structure of PRV A-capid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Sun, H. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
3SKO
 
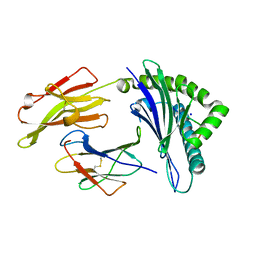 | | Crystal structure of the HLA-B8-A66-FLR, mutant A66 of the HLA B8 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
3OFS
 
 | | Dynamic conformations of the CD38-mediated NAD cyclization captured using multi-copy crystallography | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2010-08-16 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Conformations of the CD38-Mediated NAD Cyclization Captured in a Single Crystal
J.Mol.Biol., 405, 2011
|
|
3SKM
 
 | | Crystal structure of the HLA-B8FLRGRAYVL, mutant G8V of the FLR peptide | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
