4RL0
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
1T0Z
 
 | | Structure of an Excitatory Insect-specific Toxin with an Analgesic Effect on Mammalian from Scorpion Buthus martensii Karsch | | Descriptor: | SULFATE ION, insect neurotoxin | | Authors: | Li, C, Guan, R.-J, Xiang, Y, Zhang, Y, Wang, D.-C. | | Deposit date: | 2004-04-14 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an excitatory insect-specific toxin with an analgesic effect on mammals from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5YPI
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI1 complex | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPK
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI2 complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPN
 
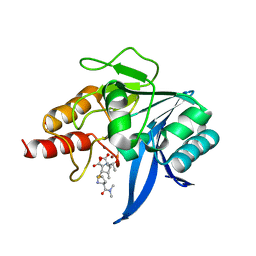 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI2 complex | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Liu, W, Wang, D. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPL
 
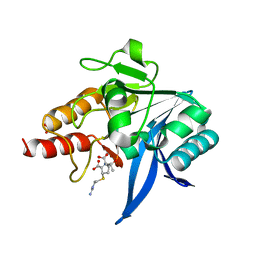 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EP complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPM
 
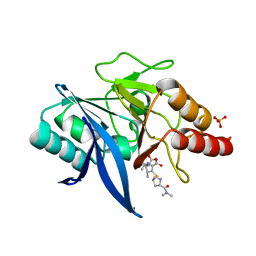 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI1 complex | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
3M3O
 
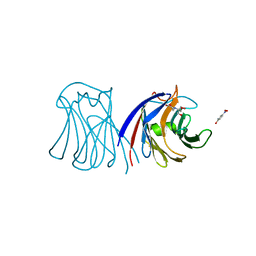 | | Crystal Structure of Agrocybe aegerita lectin AAL mutant R85A complexed with p-Nitrophenyl TF disaccharide | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-tumor lectin, P-NITROPHENOL, ... | | Authors: | Feng, L, Li, D, Wang, D. | | Deposit date: | 2010-03-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the recognition mechanism between an antitumor galectin AAL and the Thomsen-Friedenreich antigen
Faseb J., 24, 2010
|
|
3M3C
 
 | | Crystal Structure of Agrocybe aegerita lectin AAL complexed with p-Nitrophenyl TF disaccharide | | Descriptor: | Anti-tumor lectin, P-NITROPHENOL, SULFATE ION, ... | | Authors: | Feng, L, Li, D, Wang, D. | | Deposit date: | 2010-03-09 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the recognition mechanism between an antitumor galectin AAL and the Thomsen-Friedenreich antigen
Faseb J., 24, 2010
|
|
3M3E
 
 | |
3M3Q
 
 | |
7DCJ
 
 | |
7DCT
 
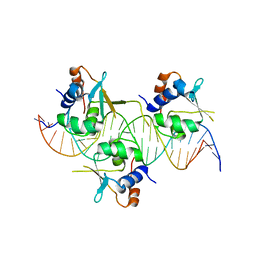 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (24 bp) | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*AP*G)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DCU
 
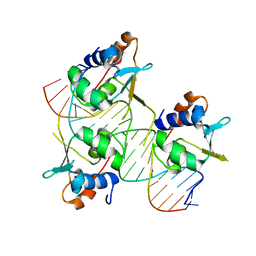 | | Crystal structure of HSF2 DNA-binding domain in complex with 3-site HSE DNA (21 bp) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*G)-3'), Heat shock factor protein 2, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DCS
 
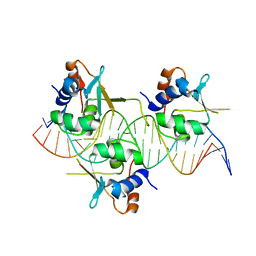 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (23 bp) | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*GP*A)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
1WAV
 
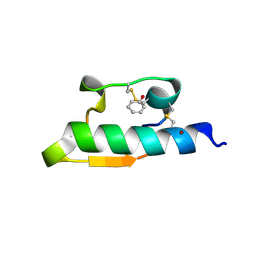 | | CRYSTAL STRUCTURE OF FORM B MONOCLINIC CRYSTAL OF INSULIN | | Descriptor: | INSULIN, PHENOL, ZINC ION | | Authors: | Liang, D.-C, Ding, J.-H, Chang, W.-R, Wan, Z.-L. | | Deposit date: | 1996-02-28 | | Release date: | 1997-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular replacement study on form-B monoclinic crystal of insulin.
Sci.China, Ser.C: Life Sci., 39, 1996
|
|
2K5Q
 
 | | NMR Solution structure of membrane associated protein from Bacillus cereus: Northeast Structural Genomics Consortium Target: BcR97A | | Descriptor: | Hypothetical Membrane Associated Protein BcR97A | | Authors: | Swapna, G, Wang, D, Owens, L, Xiao, R, Liu, J, Baran, M.C, Everett, J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution Structure of Membrane associated protein from Bacillus cereus: Northeast Structural Genomics Consortium Target: BcR97A
To be Published
|
|
5EBD
 
 | | Crystal structure of EccB1 of Mycobacterium tuberculosis in spacegroup P21 (state IV) | | Descriptor: | CALCIUM ION, CHLORIDE ION, ESX-1 secretion system protein eccB1 | | Authors: | Zhang, X.L, Qi, C, Xie, X.Q, Li, D.F, Bi, L.J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic observation of the movement of the membrane-distal domain of the T7SS core component EccB1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5EBC
 
 | | Crystal structure of EccB1 of Mycobacterium tuberculosis in spacegroup P21 (state III) | | Descriptor: | CALCIUM ION, ESX-1 secretion system protein eccB1 | | Authors: | Zhang, X.L, Qi, C, Xie, X.Q, Li, D.F, Bi, L.J. | | Deposit date: | 2015-10-19 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic observation of the movement of the membrane-distal domain of the T7SS core component EccB1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6ITS
 
 | |
6E66
 
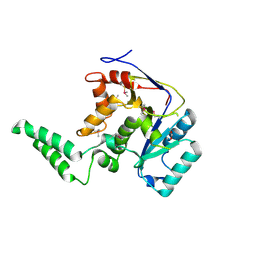 | |
3WLD
 
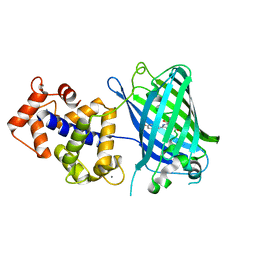 | | Crystal structure of monomeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
3WLC
 
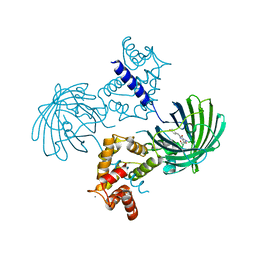 | | Crystal structure of dimeric GCaMP6m | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Ding, J, Luo, A.F, Hu, L.Y, Wang, D.C, Shao, F. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of the ultrasensitive calcium indicator GCaMP6.
Sci China Life Sci, 57, 2014
|
|
3A9U
 
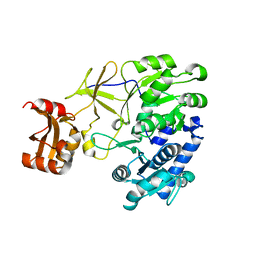 | |
3A9V
 
 | |
