3NAM
 
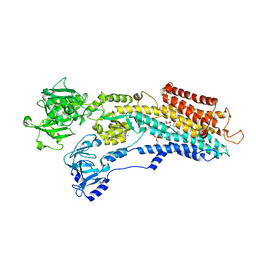 | | SR Ca(2+)-ATPase in the HnE2 state complexed with the Thapsigargin derivative dOTg | | Descriptor: | (3S,3aR,4S,6S,6aS,8R,9R,9aR,9bS)-6-(acetyloxy)-4-(butanoyloxy)-3,3a-dihydroxy-3,6,9-trimethyl-2-oxododecahydroazuleno[4,5-b]furan-8-yl (2Z)-2-methylbut-2-enoate, MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Winther, A.M.L, Sonntag, Y, Olesen, C, Moller, J.V, Nissen, P. | | Deposit date: | 2010-06-02 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Critical roles of hydrophobicity and orientation of side chains for inactivation of sarcoplasmic reticulum Ca2+-ATPase with thapsigargin and thapsigargin analogs
J.Biol.Chem., 285, 2010
|
|
1QBM
 
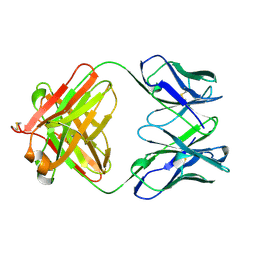 | |
4I3L
 
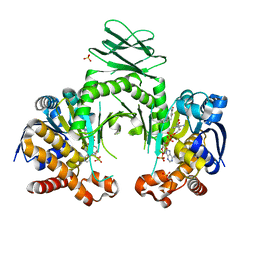 | | Crystal structure of a metabolic reductase with 6-benzyl-1-hydroxy-4-methylpyridin-2(1H)-one | | Descriptor: | 6-benzyl-1-hydroxy-4-methylpyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, B, Yao, Y, Liu, Z, Deng, L, Anglin, J.L, Jiang, H, Prasad, B.V.V, Song, Y. | | Deposit date: | 2012-11-26 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Crystallographic Investigation and Selective Inhibition of Mutant Isocitrate Dehydrogenase.
ACS Med Chem Lett, 4, 2013
|
|
4I3K
 
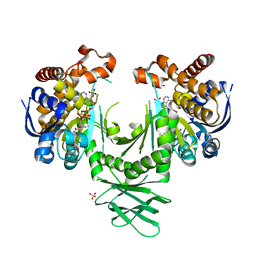 | | Crystal structure of a metabolic reductase with 1-hydroxy-6-(4-hydroxybenzyl)-4-methylpyridin-2(1H)-one | | Descriptor: | 1-hydroxy-6-(4-hydroxybenzyl)-4-methylpyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, B, Yao, Y, Liu, Z, Deng, L, Anglin, J.L, Jiang, H, Prasad, B.V.V, Song, Y. | | Deposit date: | 2012-11-26 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3056 Å) | | Cite: | Crystallographic Investigation and Selective Inhibition of Mutant Isocitrate Dehydrogenase.
ACS Med Chem Lett, 4, 2013
|
|
3RAS
 
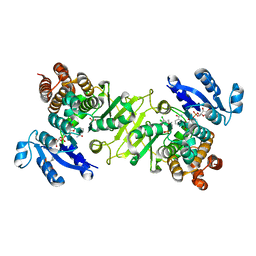 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with a lipophilic phosphonate inhibitor | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-(N-HYDROXYACETAMIDO)-1-(3,4-DICHLOROPHENYL)PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Diao, J, Deng, L, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-03-28 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase by lipophilic phosphonates: SAR, QSAR, and crystallographic studies.
J.Med.Chem., 54, 2011
|
|
1Q5K
 
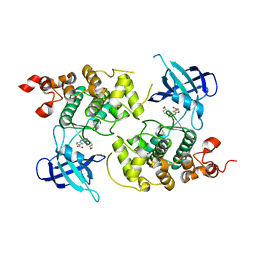 | | crystal structure of Glycogen synthase kinase 3 in complexed with inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-(4-METHOXYBENZYL)-N'-(5-NITRO-1,3-THIAZOL-2-YL)UREA | | Authors: | Bhat, R, Xue, Y, Berg, S, Hellberg, S, Ormo, M, Nilsson, Y, Radesater, A.C, Jerning, E, Markgren, P.O, Borgegard, T, Nylof, M, Gimenez-Cassina, A, Hernandez, F, Lucas, J.J, Diaz-Mido, J, Avila, J. | | Deposit date: | 2003-08-08 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418.
J.Biol.Chem., 278, 2003
|
|
3SR4
 
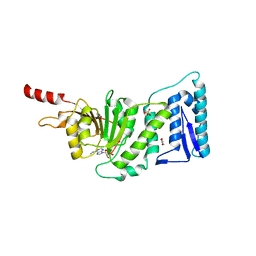 | | Crystal Structure of Human DOT1L in Complex with a Selective Inhibitor | | Descriptor: | (2S)-2-azanyl-4-[[(2S,3S,4R,5R)-5-[6-(methylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Diao, J, Chen, P, Yao, Y, Prasad, B.V.V, Song, Y. | | Deposit date: | 2011-07-06 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective Inhibitors of Histone Methyltransferase DOT1L: Design, Synthesis, and Crystallographic Studies.
J.Am.Chem.Soc., 133, 2011
|
|
1WEJ
 
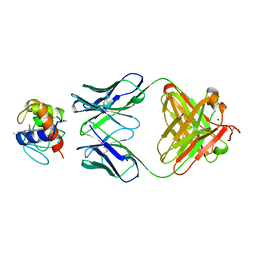 | | IGG1 FAB FRAGMENT (OF E8 ANTIBODY) COMPLEXED WITH HORSE CYTOCHROME C AT 1.8 A RESOLUTION | | Descriptor: | CYTOCHROME C, E8 ANTIBODY, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mylvaganam, S.E, Paterson, Y, Getzoff, E.D. | | Deposit date: | 1998-03-26 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the binding of an anti-cytochrome c antibody to its antigen: crystal structures of FabE8-cytochrome c complex to 1.8 A resolution and FabE8 to 2.26 A resolution.
J.Mol.Biol., 281, 1998
|
|
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
4GAE
 
 | | Crystal structure of plasmodium dxr in complex with a pyridine-containing inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, ... | | Authors: | Diao, J, Xue, J, Cai, G, Deng, L, Song, Y. | | Deposit date: | 2012-07-25 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antimalarial and Structural Studies of Pyridine-containing Inhibitors of 1-Deoxyxylulose-5-phosphate Reductoisomerase.
ACS Med Chem Lett, 4, 2013
|
|
4IN1
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, SULFATE ION | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IMQ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, PEPTIDE INHIBITOR, syc8, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IN2
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | C-like protease | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4INH
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | DIMETHYL SULFOXIDE, Genome polyprotein, peptide inhibitor, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-04 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
4IMZ
 
 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | Genome polyprotein, SODIUM ION, THIOCYANATE ION, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2013-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
3US9
 
 | | Crystal Structure of the NCX1 Intracellular Tandem Calcium Binding Domains(CBD12) | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Giladi, M, Sasson, Y, Hirsch, J.A, Khananshvili, D. | | Deposit date: | 2011-11-23 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A common Ca2+-driven interdomain module governs eukaryotic NCX regulation.
Plos One, 7, 2012
|
|
2C4J
 
 | | Human glutathione-S-transferase M2-2 T210S mutant in complex with glutathione-styrene oxide conjugate | | Descriptor: | GLUTATHIONE S-TRANSFERASE MU 2, L-GAMMA-GLUTAMYL-S-[(2S)-2-HYDROXY-2-PHENYLETHYL]-L-CYSTEINYLGLYCINE | | Authors: | Tars, K, Andersson, M, Ivarsson, Y, Olin, B, Mannervik, B. | | Deposit date: | 2005-10-20 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Alternative Mutations of a Positively Selected Residue Elicit Gain or Loss of Functionalities in Enzyme Evolution.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2IZ3
 
 | | Solution structure of human beta-microseminoprotein | | Descriptor: | BETA-MICROSEMINOPROTEIN | | Authors: | Ghasriani, H, Teilum, K, Johnsson, Y, Fernlund, P, Drakenberg, T. | | Deposit date: | 2006-07-24 | | Release date: | 2006-10-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human and Porcine Beta-Microseminoprotein
J.Mol.Biol., 362, 2006
|
|
2JCS
 
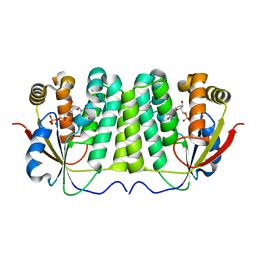 | | Active site mutant of dNK from D. melanogaster with dTTP bound | | Descriptor: | DEOXYNUCLEOSIDE KINASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Egeblad-Welin, L, Sonntag, Y, Eklund, H, Munch-Petersen, B. | | Deposit date: | 2007-01-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Studies of Active-Site Mutants from Drosophila Melanogaster Deoxyribonucleoside Kinase. Investigations of the Putative Catalytic Glutamate- Arginine Pair and of Residues Responsible for Substrate Specificity.
FEBS J., 274, 2007
|
|
2IZ4
 
 | | Solution structure of porcine beta-microseminoprotein | | Descriptor: | BETA-MICROSEMINOPROTEIN | | Authors: | Ghasriani, H, Teilum, K, Johnsson, Y, Fernlund, P, Drakenberg, T. | | Deposit date: | 2006-07-24 | | Release date: | 2006-10-24 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human and Porcine Beta-Microseminoprotein
J.Mol.Biol., 362, 2006
|
|
2LP1
 
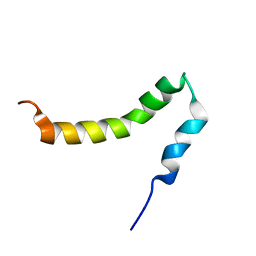 | | The solution NMR structure of the transmembrane C-terminal domain of the amyloid precursor protein (C99) | | Descriptor: | C99 | | Authors: | Barrett, P.J, Song, Y, Van Horn, W.D, Hustedt, E.J, Schafer, J.M, Hadziselimovic, A, Beel, A.J, Sanders, C.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol.
Science, 336, 2012
|
|
2MV0
 
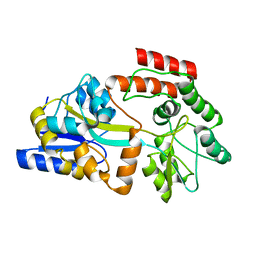 | | Solution NMR Structure of Maltose-binding protein from Escherichia coli, Northeast Structural Genomics Consortium (NESG) Target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Rossi, P, Lange, O.F, Sgourakis, N.G, Song, Y, Lee, H, Aramini, J.M, Ertekin, A, Xiao, R, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2NDJ
 
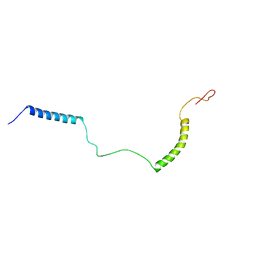 | | Structural Basis for KCNE3 and Estrogen Modulation of the KCNQ1 Channel | | Descriptor: | Potassium voltage-gated channel subfamily E member 3 | | Authors: | Sanders, C.R, Van Horn, W.D, Kroncke, B.M, Sisco, N.J, Meiler, J, Vanoye, C.G, Song, Y, Nannemann, D.P, Welch, R.C, Kang, C, Smith, J, George, A.L. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for KCNE3 modulation of potassium recycling in epithelia.
Sci Adv, 2, 2016
|
|
7CSW
 
 | | Pseudomonas aeruginosa antitoxin HigA with pa2440 promoter | | Descriptor: | HTH cro/C1-type domain-containing protein, pa2440 | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
