6ZYQ
 
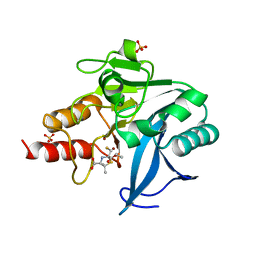 | | Structure of NDM-1 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
5HH5
 
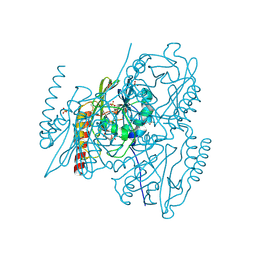 | |
5ACR
 
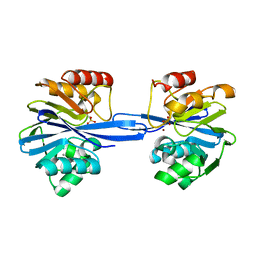 | | W228Y-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | CALCIUM ION, GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
7AJN
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with a BzD ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(1-adamantylmethyl)-2-[(7~{R},9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,10,12-tetraen-9-yl]ethanamide | | Authors: | Picaud, S, Hassel-Hart, S, Tobias, K, Spencer, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of the first bromodomain of BRD4 in complex with a BzD ligand
To Be Published
|
|
6SE4
 
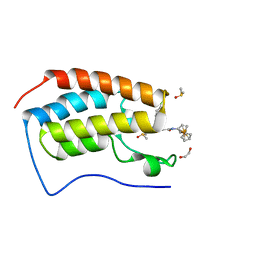 | | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor | | Descriptor: | (+)-JD1, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Hassell-Hart, S, Picaud, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Spencer, J, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor
To Be Published
|
|
5ACT
 
 | | W228S-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
6ZYS
 
 | | Structure of IMP-1 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase IMP-1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87000954 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
6ZYN
 
 | | Structure of VIM-2 with 2-Mercaptomethyl-thiazolidine L-anti-1b | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4000138 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
6ZYP
 
 | | Structure of NDM-1 with 2-Mercaptomethyl-thiazolidine L-anti-1b | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
5ACQ
 
 | | W228A-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | BETA-LACTAMASE, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
7BJ9
 
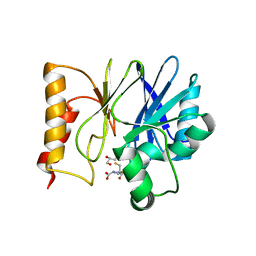 | | Structure of Sfh-I with 2-Mercaptomethyl-thiazolidine L-anti-1a | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase, GLYCEROL, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21000588 Å) | | Cite: | 2-Mercaptomethyl Thiazolidines (MMTZs) Inhibit All Metallo-beta-Lactamase Classes by Maintaining a Conserved Binding Mode.
Acs Infect Dis., 7, 2021
|
|
7BJ8
 
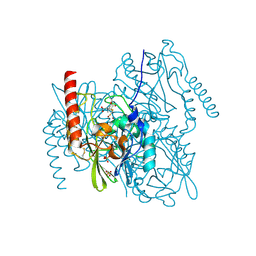 | | Structure of L1 with 2-Mercaptomethyl-thiazolidine D-syn-1b | | Descriptor: | (2~{S},4~{S})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | 2-Mercaptomethyl Thiazolidines (MMTZs) Inhibit All Metallo-beta-Lactamase Classes by Maintaining a Conserved Binding Mode.
Acs Infect Dis., 7, 2021
|
|
5ACS
 
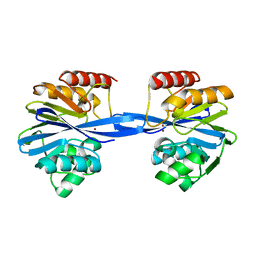 | | Y233A-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
7ZO5
 
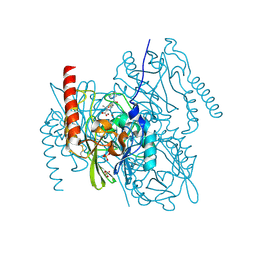 | | L1 metallo-beta-lactamase in complex with a mecillinam degradation product | | Descriptor: | (2~{R},4~{S})-2-[(1~{R})-2-(azepan-1-yl)-1-formamido-2-oxidanylidene-ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO2
 
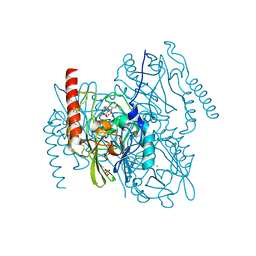 | | L1 metallo-beta-lactamase complex with hydrolysed doripenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase L1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO3
 
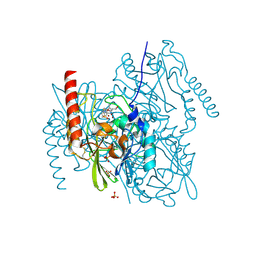 | | L1 metallo-beta-lactamase in complex with hydrolysed tebipenem | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[1-(4,5-dihydro-1,3-thiazol-2-yl)azetidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO4
 
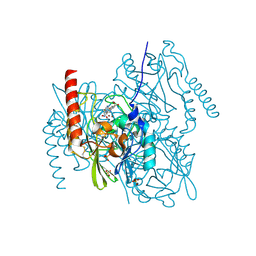 | | L1 metallo-beta-lactamase in complex with hydrolysed panipenem | | Descriptor: | (2R,4S)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S)-1-ethanimidoylpyrrolidin-3-yl]sulfanyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SODIUM ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO6
 
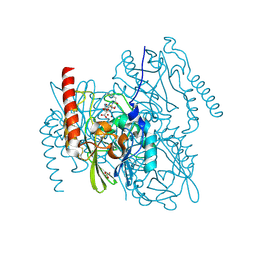 | | L1 metallo-beta-lactamase in complex with hydrolysed cefoxitin | | Descriptor: | (2~{R},5~{S})-5-(aminocarbonyloxymethyl)-2-[(1~{S})-1-methoxy-2-oxidanyl-2-oxidanylidene-1-(2-thiophen-2-ylethanoylamino)ethyl]-5,6-dihydro-2~{H}-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7ZO7
 
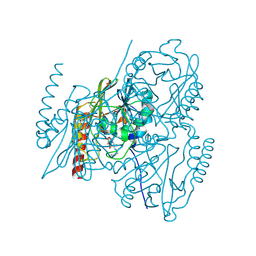 | | L1 metallo-beta-lactamase in complex with hydrolysed cefmetazole | | Descriptor: | (2R,5R)-2-[(1S)-1-[2-(cyanomethylsulfanyl)ethanoylamino]-1-methoxy-2-oxidanyl-2-oxidanylidene-ethyl]-5-[(1-methyl-1,2,3,4-tetrazol-5-yl)sulfanylmethyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-04-24 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interactions of hydrolyzed beta-lactams with the L1 metallo-beta-lactamase: Crystallography supports stereoselective binding of cephem/carbapenem products.
J.Biol.Chem., 299, 2023
|
|
7BDR
 
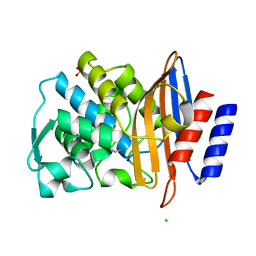 | | Structure of CTX-M-15 E166Q mutant crystallised in the presence of tazobactam (AAI101) | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-12-22 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Penicillanic Acid Sulfones Inactivate the Extended-Spectrum beta-Lactamase CTX-M-15 through Formation of a Serine-Lysine Cross-Link: an Alternative Mechanism of beta-Lactamase Inhibition.
Mbio, 13, 2022
|
|
7BDS
 
 | |
6SUT
 
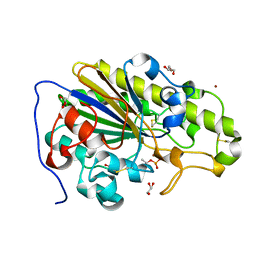 | | Crystal structure of phosphothreonine MCR-2 | | Descriptor: | BROMIDE ION, GLYCEROL, Putative integral membrane protein, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-09-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Resistance to the "last resort" antibiotic colistin: a single-zinc mechanism for phosphointermediate formation in MCR enzymes.
Chem.Commun.(Camb.), 56, 2020
|
|
8AKK
 
 | | Acyl-enzyme complex of imipenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKI
 
 | | Acyl-enzyme complex of ampicillin bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKM
 
 | | Acyl-enzyme complex of ertapenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, Ertapenem, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
