3IMO
 
 | | Structure from the mobile metagenome of Vibrio cholerae. Integron cassette protein VCH_CASS14 | | Descriptor: | ACETATE ION, Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2009-08-11 | | Release date: | 2009-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites
Plos One, 8, 2013
|
|
3J5P
 
 | |
7C5D
 
 | | Crystal structure of TRF2 TRFH domain in complex with a MCPH1 peptide | | Descriptor: | GLYCEROL, Microcephalin, Telomeric repeat-binding factor 2 | | Authors: | Xiong, X, Chen, Y. | | Deposit date: | 2020-05-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Microcephalin 1/BRIT1-TRF2 interaction promotes telomere replication and repair, linking telomere dysfunction to primary microcephaly.
Nat Commun, 11, 2020
|
|
7E1A
 
 | |
7EHW
 
 | | BRD4-BD1 in complex with LT-642-602 | | Descriptor: | 1-[4-ethyl-2-methyl-5-(6-morpholin-4-yl-1H-benzimidazol-2-yl)-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | BRD4-BD1 in complex with LT-642-602
To Be Published
|
|
7EHY
 
 | | BRD4-BD1 in complex with LT-448-138 | | Descriptor: | 1-[2,4-dimethyl-5-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | BRD4-BD1 in complex with LT-642-602
To Be Published
|
|
7EIG
 
 | | BRD4-BD1 in complex with LT-730-903 | | Descriptor: | 1-[5-(5-azanyl-1H-benzimidazol-2-yl)-2-methyl-4-phenyl-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | BRD4-BD1 in complex with LT-730-903
To Be Published
|
|
7EIK
 
 | | RD4-BD1 in complex with LT-872-297 | | Descriptor: | 1-[2-methyl-5-[6-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-4-(1-methylpyrazol-4-yl)-1H-pyrrol-3-yl]ethanone, Bromodomain-containing protein 4 | | Authors: | Zheng, W, Kong, B, Tang, W, Zhu, J, Chen, Y. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RD4-BD1 in complex with LT-872-297
To Be Published
|
|
7D4O
 
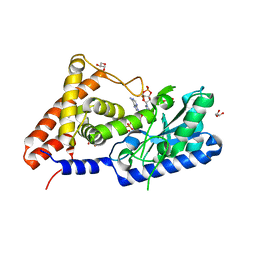 | | cyclic trinucleotide synthase CdnD in complex with ATP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase, ... | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4J
 
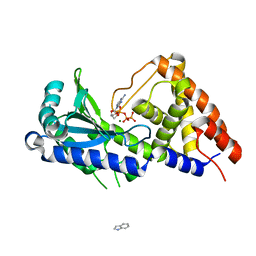 | | ddATP complex of cyclic trinucleotide synthase CdnD | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, Cyclic AMP-AMP-GMP synthase, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D48
 
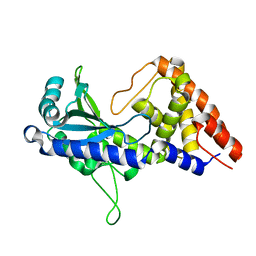 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, SODIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4U
 
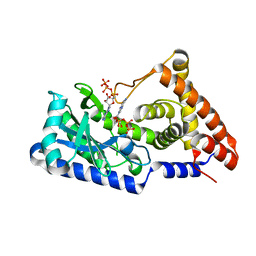 | | ATP complex with double mutant cyclic trinucleotide synthase CdnD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7D4S
 
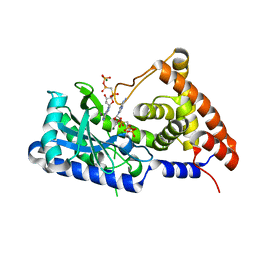 | | apo-form cyclic trinucleotide synthase CdnD | | Descriptor: | Cyclic AMP-AMP-GMP synthase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION | | Authors: | Yang, C.-S, Hou, M.-H, Tsai, C.-L, Wang, Y.-C, Ko, T.-P, Chen, Y. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure and functional implication of a bacterial cyclic AMP-AMP-GMP synthetase.
Nucleic Acids Res., 49, 2021
|
|
7DV5
 
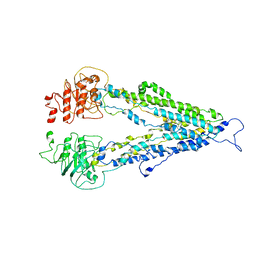 | |
7DD0
 
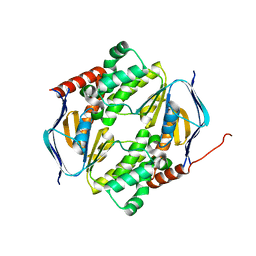 | |
7F8E
 
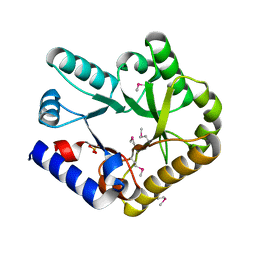 | |
7CAM
 
 | | SARS-CoV-2 main protease (Mpro) apo structure (space group P212121) | | Descriptor: | 3C-like proteinase | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y. | | Deposit date: | 2020-06-09 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
7CB7
 
 | | 1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y, Hung, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
7FBI
 
 | | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5 | | Descriptor: | 3A3 heavy chain, 3A3 light chain, 3A5 heavy chain, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Hong, J, Chen, Y, Zhang, X. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of EBV gB in complex with nAbs 3A3 and 3A5
To Be Published
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
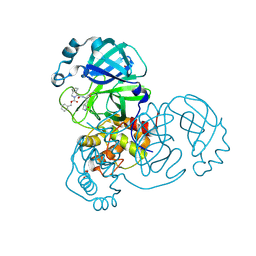
7LYI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-3 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7LYH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7K99
 
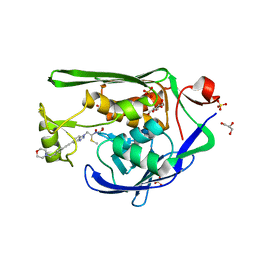 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor 19 | | Descriptor: | (hydroxy{(1S)-1-(methylsulfanyl)-2-[5-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)-1H-benzotriazol-1-yl]ethyl}amino)methanol, GLYCEROL, SULFATE ION, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
7K9A
 
 | | Crystal Structure of P. aeruginosa LpxC with N-Hydroxyformamide inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-hydroxy-N-[(1R)-2-{5-[(4-{[2-(hydroxymethyl)-1H-imidazol-1-yl]methyl}phenyl)ethynyl]-1H-benzotriazol-1-yl}-1-(methylsulfanyl)ethyl]formamide, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-Hydroxyformamide LpxC inhibitors, their in vivo efficacy in a mouse Escherichia coli infection model, and their safety in a rat hemodynamic assay.
Bioorg.Med.Chem., 28, 2020
|
|
7K65
 
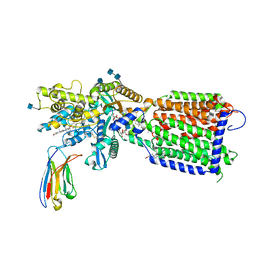 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
