5Y0J
 
 | | Solution structure of arenicin-3 derivative N2 | | Descriptor: | N2 | | Authors: | Liu, X.H, Wang, J.H. | | Deposit date: | 2017-07-17 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis
Sci Rep, 7, 2017
|
|
4RB4
 
 | | Crystal structure of dodecameric iron-containing heptosyltransferase TibC in complex with ADP-D-beta-D-heptose at 3.9 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Glycosyltransferase tibC, ... | | Authors: | Yao, Q, Lu, Q, Shao, F. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family
Elife, 3, 2014
|
|
7CIR
 
 | | Peptide phosphorylation modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIQ
 
 | | Phosphorylation modification of MHC I polypeptide | | Descriptor: | ARG-ARG-PHE-SER-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIS
 
 | | Peptide modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CU6
 
 | | lasso peptide C24 mutant - A11V2C | | Descriptor: | lasso peptide C24_A11V2C | | Authors: | Ma, M, Liu, X.H. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
5WYM
 
 | | Crystal structure of an anti-connexin26 scFv | | Descriptor: | anti-connexin26 scFv,Ig heavy chain,Linker,anti-connexin26 scFv,Ig light chain | | Authors: | Li, S, Xu, L. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Characterization of a Human Monoclonal Antibody that Modulates Mutant Connexin 26 Hemichannels Implicated in Deafness and Skin Disorders
Front Mol Neurosci, 10, 2017
|
|
7DYN
 
 | | Phosphorylation of MHC I peptide | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
5YPI
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI1 complex | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5UTT
 
 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5YPK
 
 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EI2 complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
7YEU
 
 | | Superfolder green fluorescent protein with phosphine unnatural amino acid P3BF | | Descriptor: | Superfolder green fluorescent protein | | Authors: | Hu, C, Duan, H.Z, Liu, X.H, Chen, Y.X, Wang, J.Y. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically Encoded Phosphine Ligand for Metalloprotein Design.
J.Am.Chem.Soc., 144, 2022
|
|
5YPN
 
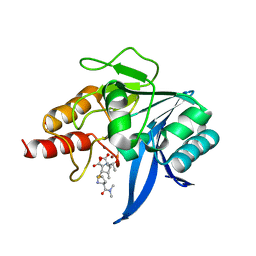 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI2 complex | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Liu, W, Wang, D. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPL
 
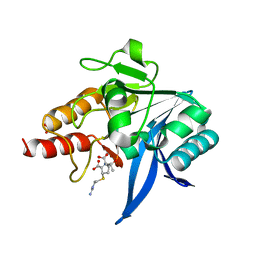 | | Crystal structure of NDM-1 bound to hydrolyzed imipenem representing an EP complex | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
5YPM
 
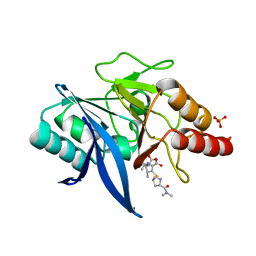 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI1 complex | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
8V17
 
 | |
8W56
 
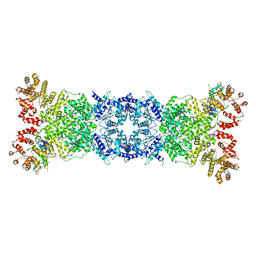 | | Cryo-EM structure of DSR2-DSAD1 state 1 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-08-25 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
7ZRB
 
 | |
8Y87
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y88
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y89
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Wang, H.F, Zhang, X, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8B
 
 | | Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, H.F, Zhang, X, Lu, Y.C, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y7X
 
 | | Structure of HCoV-HKU1A spike in the functionally anchored-3up conformation with 3TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8A
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 3TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8WKN
 
 | | Cryo-EM structure of DSR2-DSAD1 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-09-28 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
